Insights from the analysis of draft genome sequence of Crocus sativus L
- PMID: 35815202
- PMCID: PMC9200609
- DOI: 10.6026/97320630018001
Insights from the analysis of draft genome sequence of Crocus sativus L
Abstract
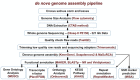
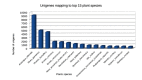
Saffron (Crocus sativus L.) is the low yielding plant of medicinal and economic importance. Therefore, it is of interest to report the draft genome sequence of C. sativus. The draft genome of C. sativus has been assembled using Illumina sequencing and is 3.01 Gb long covering 84.24% of genome. C. sativus genome annotation identified 53,546 functional genes (including 5726 transcription factors), 862,275 repeats and 964,231 SSR markers. The genes involved in the apocarotenoids biosynthesis pathway (crocin, crocetin, picrocrocin, and safranal) were found in the draft genome analysis.
Keywords: Crocus sativus; MYB TFs; Orthology analysis; SSR markers; apocarotene biosynthesis pathway; de-novo genome assembly.
© 2022 Biomedical Informatics.
Conflict of interest statement
The authors declare no conflict of Interest.
Figures

References
-
- Liu J, et al. Genome Biol. . 2020;21:121. - PMC - PubMed
- {'text': '', 'ref_index': 2, 'ids': [{'type': 'DOI', 'value': '10.1186/s13059-020-02029-9', 'is_inner': False, 'url': 'https://doi.org/10.1186/s13059-020-02029-9'}]}
-
- Xia E, et al. Mol Plant. 2020;13:1013. - PubMed
LinkOut - more resources
Full Text Sources
