Molecular epidemiology of Porcine Parvovirus Type 1 (PPV1) and the reactivity of vaccine-induced antisera against historical and current PPV1 strains
- PMID: 35815310
- PMCID: PMC9252332
- DOI: 10.1093/ve/veac053
Molecular epidemiology of Porcine Parvovirus Type 1 (PPV1) and the reactivity of vaccine-induced antisera against historical and current PPV1 strains
Abstract
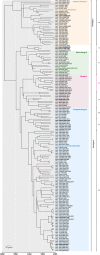
Porcine Parvovirus Type 1 (PPV1) contributes to important losses in the swine industry worldwide. During a PPV1 infection, embryos and fetuses are targeted, resulting in stillbirth, mummification, embryonic death, and infertility (SMEDI syndrome). Even though vaccination is common in gilts and sows, strains mainly belonging to the 27a-like group have been spreading in Europe since early 2000s, resulting in SMEDI problems and requiring in-depth studies into the molecular epidemiology and vaccination efficacy of commercial vaccines. Here, we show that PPV1 has evolved since 1855 [1737, 1933] at a rate of 4.71 × 10-5 nucleotide substitutions per site per year. Extensive sequencing allowed evaluating and reassessing the current PPV1 VP1-based classifications, providing evidence for the existence of four relevant phylogenetic groups. While most European strains belong to the PPV1a (G1) or PPV1b (G2 or 27a-like) group, most Asian and American G2 strains and some European strains were divided into virulent PPV1c (e.g. NADL-8) and attenuated PPV1d (e.g. NADL-2) groups. The increase in the swine population, vaccination degree, and health management (vaccination and biosafety) influenced the spread of PPV1. The reactivity of anti-PPV1 antibodies from sows vaccinated with Porcilis© Parvo, Eryseng© Parvo, or ReproCyc© ParvoFLEX against different PPV1 field strains was the highest upon vaccination with ReproCyc© ParvoFLEX, followed by Eryseng© Parvo, and Porcilis© Parvo. Our findings contribute to the evaluation of the immunogenicity of existing vaccines and support the development of new vaccine candidates. Finally, the potential roles of cluster-specific hallmark amino acids in elevated pathogenicity and viral entry are discussed.
Keywords: BEAST; evolution; molecular phylodynamics; vaccination; viral classification.
© The Author(s) 2022. Published by Oxford University Press.
Figures




References
LinkOut - more resources
Full Text Sources

