A machine learning approach using partitioning around medoids clustering and random forest classification to model groups of farms in regard to production parameters and bulk tank milk antibody status of two major internal parasites in dairy cows
- PMID: 35816512
- PMCID: PMC9273072
- DOI: 10.1371/journal.pone.0271413
A machine learning approach using partitioning around medoids clustering and random forest classification to model groups of farms in regard to production parameters and bulk tank milk antibody status of two major internal parasites in dairy cows
Abstract
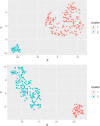
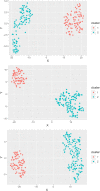
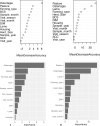
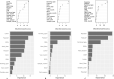
Fasciola hepatica and Ostertagia ostertagi are internal parasites of cattle compromising physiology, productivity, and well-being. Parasites are complex in their effect on hosts, sometimes making it difficult to identify clear directions of associations between infection and production parameters. Therefore, unsupervised approaches not assuming a structure reduce the risk of introducing bias to the analysis. They may provide insights which cannot be obtained with conventional, supervised methodology. An unsupervised, exploratory cluster analysis approach using the k-mode algorithm and partitioning around medoids detected two distinct clusters in a cross-sectional data set of milk yield, milk fat content, milk protein content as well as F. hepatica or O. ostertagi bulk tank milk antibody status from 606 dairy farms in three structurally different dairying regions in Germany. Parasite-positive farms grouped together with their respective production parameters to form separate clusters. A random forests algorithm characterised clusters with regard to external variables. Across all study regions, co-infections with F. hepatica or O. ostertagi, respectively, farming type, and pasture access appeared to be the most important factors discriminating clusters (i.e. farms). Furthermore, farm level lameness prevalence, herd size, BCS, stage of lactation, and somatic cell count were relevant criteria distinguishing clusters. This study is among the first to apply a cluster analysis approach in this context and potentially the first to implement a k-medoids algorithm and partitioning around medoids in the veterinary field. The results demonstrated that biologically relevant patterns of parasite status and milk parameters exist between farms positive for F. hepatica or O. ostertagi, respectively, and negative farms. Moreover, the machine learning approach confirmed results of previous work and shed further light on the complex setting of associations a between parasitic diseases, milk yield and milk constituents, and management practices.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Breed-dependent associations of production characteristics with on-farm seropositivity for Ostertagia ostertagi in dairy cows.Parasit Vectors. 2025 Mar 5;18(1):91. doi: 10.1186/s13071-025-06728-9. Parasit Vectors. 2025. PMID: 40045409 Free PMC article.
-
Random forest classification as a tool in epidemiological modelling: Identification of farm-specific characteristics relevant for the occurrence of Fasciola hepatica on German dairy farms.PLoS One. 2023 Dec 21;18(12):e0296093. doi: 10.1371/journal.pone.0296093. eCollection 2023. PLoS One. 2023. PMID: 38128054 Free PMC article.
-
Multinomial logistic regression based on neural networks reveals inherent differences among dairy farms depending on the differential exposure to Fasciola hepatica and Ostertagia ostertagi.Int J Parasitol. 2023 Oct;53(11-12):687-697. doi: 10.1016/j.ijpara.2023.05.006. Epub 2023 Jun 22. Int J Parasitol. 2023. PMID: 37355196
-
Milk as a diagnostic fluid to monitor viral diseases in dairy cattle.Aust Vet J. 2024 Jan-Feb;102(1-2):11-18. doi: 10.1111/avj.13293. Epub 2023 Oct 9. Aust Vet J. 2024. PMID: 37814548 Review.
-
Over 20 Years of Machine Learning Applications on Dairy Farms: A Comprehensive Mapping Study.Sensors (Basel). 2021 Dec 22;22(1):52. doi: 10.3390/s22010052. Sensors (Basel). 2021. PMID: 35009593 Free PMC article. Review.
Cited by
-
Breed-dependent associations of production characteristics with on-farm seropositivity for Ostertagia ostertagi in dairy cows.Parasit Vectors. 2025 Mar 5;18(1):91. doi: 10.1186/s13071-025-06728-9. Parasit Vectors. 2025. PMID: 40045409 Free PMC article.
-
Animal health as a function of farmer personality and attitude: using the HEXACO model of personality structure to predict farm-level seropositivity for Fasciola hepatica and Ostertagia ostertagi in dairy cows.Front Vet Sci. 2024 Oct 2;11:1434612. doi: 10.3389/fvets.2024.1434612. eCollection 2024. Front Vet Sci. 2024. PMID: 39415948 Free PMC article.
-
Associations of production characteristics with the on-farm presence of Fasciola hepatica in dairy cows vary across production levels and indicate differences between breeds.PLoS One. 2023 Nov 17;18(11):e0294601. doi: 10.1371/journal.pone.0294601. eCollection 2023. PLoS One. 2023. PMID: 37976265 Free PMC article.
-
Random forest classification as a tool in epidemiological modelling: Identification of farm-specific characteristics relevant for the occurrence of Fasciola hepatica on German dairy farms.PLoS One. 2023 Dec 21;18(12):e0296093. doi: 10.1371/journal.pone.0296093. eCollection 2023. PLoS One. 2023. PMID: 38128054 Free PMC article.
-
Establishment and Validation of Fourier Transform Infrared Spectroscopy (FT-MIR) Methodology for the Detection of Linoleic Acid in Buffalo Milk.Foods. 2023 Mar 12;12(6):1199. doi: 10.3390/foods12061199. Foods. 2023. PMID: 36981127 Free PMC article.
References
-
- Chaparro JJ, Ramírez NF, Villar D, Fernandez JA, Londoño J, Arbeláez C, et al.. Survey of gastrointestinal parasites, liver flukes and lungworm in feces from dairy cattle in the high tropics of Antioquia, Colombia. Parasite Epidemiol Control. 2016;1:124–30. doi: 10.1016/j.parepi.2016.05.001 - DOI - PMC - PubMed
-
- Bellet C, Green MJ, Vickers M, Forbes A, Berry E, Kaler J. Ostertagia spp., rumen fluke and liver fluke single- and poly-infections in cattle: An abattoir study of prevalence and production impacts in England and Wales. Prev Vet Med. 2016;132:98–106. doi: 10.1016/j.prevetmed.2016.08.010 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

