Regulation of AUXIN RESPONSE FACTOR condensation and nucleo-cytoplasmic partitioning
- PMID: 35817767
- PMCID: PMC9273615
- DOI: 10.1038/s41467-022-31628-2
Regulation of AUXIN RESPONSE FACTOR condensation and nucleo-cytoplasmic partitioning
Abstract
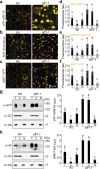
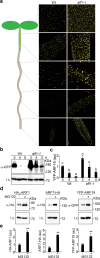
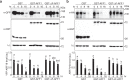
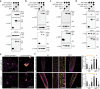
Auxin critically regulates plant growth and development. Auxin-driven transcriptional responses are mediated through the AUXIN RESPONSE FACTOR (ARF) family of transcription factors. ARF protein condensation attenuates ARF activity, resulting in dramatic shifts in the auxin transcriptional landscape. Here, we perform a forward genetics screen for ARF hypercondensation, identifying an F-box protein, which we named AUXIN RESPONSE FACTOR F-BOX1 (AFF1). Functional characterization of SCFAFF1 revealed that this E3 ubiquitin ligase directly interacts with ARF19 and ARF7 to regulate their accumulation, condensation, and nucleo-cytoplasmic partitioning. Mutants defective in AFF1 display attenuated auxin responsiveness, and developmental defects, suggesting that SCFAFF1 -mediated regulation of ARF protein drives aspects of auxin response and plant development.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

