Transition to a mesenchymal state in neuroblastoma confers resistance to anti-GD2 antibody via reduced expression of ST8SIA1
- PMID: 35817829
- PMCID: PMC10071839
- DOI: 10.1038/s43018-022-00405-x
Transition to a mesenchymal state in neuroblastoma confers resistance to anti-GD2 antibody via reduced expression of ST8SIA1
Abstract
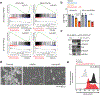
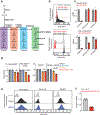
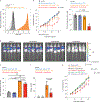
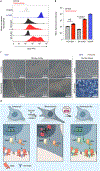
Immunotherapy with anti-GD2 antibodies has advanced the treatment of children with high-risk neuroblastoma, but nearly half of patients relapse, and little is known about mechanisms of resistance to anti-GD2 therapy. Here, we show that reduced GD2 expression was significantly correlated with the mesenchymal cell state in neuroblastoma and that a forced adrenergic-to-mesenchymal transition (AMT) conferred downregulation of GD2 and resistance to anti-GD2 antibody. Mechanistically, low-GD2-expressing cell lines demonstrated significantly reduced expression of the ganglioside synthesis enzyme ST8SIA1 (GD3 synthase), resulting in a bottlenecking of GD2 synthesis. Pharmacologic inhibition of EZH2 resulted in epigenetic rewiring of mesenchymal neuroblastoma cells and re-expression of ST8SIA1, restoring surface expression of GD2 and sensitivity to anti-GD2 antibody. These data identify developmental lineage as a key determinant of sensitivity to anti-GD2 based immunotherapies and credential EZH2 inhibitors for clinical testing in combination with anti-GD2 antibody to enhance outcomes for children with neuroblastoma.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
The remaining authors declare no competing interests.
Figures














References
Publication types
MeSH terms
Substances
Grants and funding
- UM1 HG012076/HG/NHGRI NIH HHS/United States
- K99 CA279915/CA/NCI NIH HHS/United States
- DP1 NS111132/NS/NINDS NIH HHS/United States
- F32 CA261035/CA/NCI NIH HHS/United States
- R01 NS092597/NS/NINDS NIH HHS/United States
- R01 NS088355/NS/NINDS NIH HHS/United States
- P01 CA217959/CA/NCI NIH HHS/United States
- R35 CA210030/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 CA227942/CA/NCI NIH HHS/United States
- F30 CA232541/CA/NCI NIH HHS/United States
- CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

