Directed cell migration towards softer environments
- PMID: 35817964
- PMCID: PMC10712428
- DOI: 10.1038/s41563-022-01294-2
Directed cell migration towards softer environments
Abstract
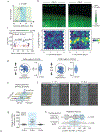
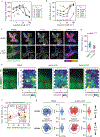
How cells sense tissue stiffness to guide cell migration is a fundamental question in development, fibrosis and cancer. Although durotaxis-cell migration towards increasing substrate stiffness-is well established, it remains unknown whether individual cells can migrate towards softer environments. Here, using microfabricated stiffness gradients, we describe the directed migration of U-251MG glioma cells towards less stiff regions. This 'negative durotaxis' does not coincide with changes in canonical mechanosensitive signalling or actomyosin contractility. Instead, as predicted by the motor-clutch-based model, migration occurs towards areas of 'optimal stiffness', where cells can generate maximal traction. In agreement with this model, negative durotaxis is selectively disrupted and even reversed by the partial inhibition of actomyosin contractility. Conversely, positive durotaxis can be switched to negative by lowering the optimal stiffness by the downregulation of talin-a key clutch component. Our results identify the molecular mechanism driving context-dependent positive or negative durotaxis, determined by a cell's contractile and adhesive machinery.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Conflicts of Interest
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

