Transcriptomics for child and adolescent tuberculosis
- PMID: 35818983
- PMCID: PMC9540430
- DOI: 10.1111/imr.13116
Transcriptomics for child and adolescent tuberculosis
Abstract
Tuberculosis (TB) in humans is caused by Mycobacterium tuberculosis (Mtb). It is estimated that 70 million children (<15 years) are currently infected with Mtb, with 1.2 million each year progressing to disease. Of these, a quarter die. The risk of progression from Mtb infection to disease and from disease to death is dependent on multiple pathogen and host factors. Age is a central component in all these transitions. The natural history of TB in children and adolescents is different to adults, leading to unique challenges in the development of diagnostics, therapeutics, and vaccines. The quantification of RNA transcripts in specific cells or in the peripheral blood, using high-throughput methods, such as microarray analysis or RNA-Sequencing, can shed light into the host immune response to Mtb during infection and disease, as well as understanding treatment response, disease severity, and vaccination, in a global hypothesis-free manner. Additionally, gene expression profiling can be used for biomarker discovery, to diagnose disease, predict future disease progression and to monitor response to treatment. Here, we review the role of transcriptomics in children and adolescents, focused mainly on work done in blood, to understand disease biology, and to discriminate disease states to assist clinical decision-making. In recent years, studies with a specific pediatric and adolescent focus have identified blood gene expression markers with diagnostic or prognostic potential that meet or exceed the current sensitivity and specificity targets for diagnostic tools. Diagnostic and prognostic gene expression signatures identified through high-throughput methods are currently being translated into diagnostic tests.
Keywords: children; diagnosis; differential expression; transcriptomics; tuberculosis.
© 2022 The Authors. Immunological Reviews published by John Wiley & Sons Ltd.
Conflict of interest statement
MK, ML, and TS hold patents on several gene expression signatures. CB, OV, and JAS have no conflicts of interest.
Figures
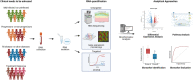
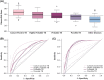
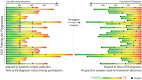


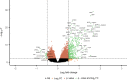
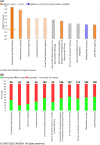
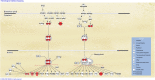
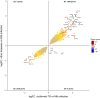
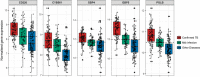
References
-
- Daniel TM. The history of tuberculosis. Respir Med. 2006;100(11):1862‐1870. - PubMed
-
- Jones C, Whittaker E, Bamford A, Kampmann B. Immunology and pathogenesis of childhood TB. Paediatr Respir Rev. 2011;12(1):3‐8. - PubMed
-
- Marais BJ, Gie RP, Schaaf HS, Hesseling AC, Enarson DA, Beyers N. The spectrum of disease in children treated for tuberculosis in a highly endemic area. Int J Tuberc Lung Dis. 2006;10(7):732‐738. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

