NURF301 contributes to gypsy chromatin insulator-mediated nuclear organization
- PMID: 35819192
- PMCID: PMC9371915
- DOI: 10.1093/nar/gkac600
NURF301 contributes to gypsy chromatin insulator-mediated nuclear organization
Abstract
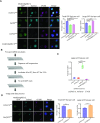
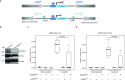
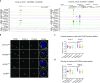
Chromatin insulators are DNA-protein complexes that can prevent the spread of repressive chromatin and block communication between enhancers and promoters to regulate gene expression. In Drosophila, the gypsy chromatin insulator complex consists of three core proteins: CP190, Su(Hw), and Mod(mdg4)67.2. These factors concentrate at nuclear foci termed insulator bodies, and changes in insulator body localization have been observed in mutants defective for insulator function. Here, we identified NURF301/E(bx), a nucleosome remodeling factor, as a novel regulator of gypsy insulator body localization through a high-throughput RNAi imaging screen. NURF301 promotes gypsy-dependent insulator barrier activity and physically interacts with gypsy insulator proteins. Using ChIP-seq, we found that NURF301 co-localizes with insulator proteins genome-wide, and NURF301 promotes chromatin association of Su(Hw) and CP190 at gypsy insulator binding sites. These effects correlate with NURF301-dependent nucleosome repositioning. At the same time, CP190 and Su(Hw) both facilitate recruitment of NURF301 to chromatin. Finally, Oligopaint FISH combined with immunofluorescence revealed that NURF301 promotes 3D contact between insulator bodies and gypsy insulator DNA binding sites, and NURF301 is required for proper nuclear positioning of gypsy binding sites. Our data provide new insights into how a nucleosome remodeling factor and insulator proteins cooperatively contribute to nuclear organization.
Published by Oxford University Press on behalf of Nucleic Acids Research 2022.
Figures




Similar articles
-
The zinc-finger protein CLAMP promotes gypsy chromatin insulator function in Drosophila.J Cell Sci. 2019 Mar 8;132(5):jcs226092. doi: 10.1242/jcs.226092. J Cell Sci. 2019. PMID: 30718365 Free PMC article.
-
The centrosomal protein CP190 is a component of the gypsy chromatin insulator.Mol Cell. 2004 Dec 3;16(5):737-48. doi: 10.1016/j.molcel.2004.11.004. Mol Cell. 2004. PMID: 15574329
-
Genome-Wide Mapping Targets of the Metazoan Chromatin Remodeling Factor NURF Reveals Nucleosome Remodeling at Enhancers, Core Promoters and Gene Insulators.PLoS Genet. 2016 Apr 5;12(4):e1005969. doi: 10.1371/journal.pgen.1005969. eCollection 2016 Apr. PLoS Genet. 2016. PMID: 27046080 Free PMC article.
-
Functional sub-division of the Drosophila genome via chromatin looping: the emerging importance of CP190.Nucleus. 2013 Mar-Apr;4(2):115-22. doi: 10.4161/nucl.23389. Epub 2013 Jan 18. Nucleus. 2013. PMID: 23333867 Free PMC article. Review.
-
Genetic and molecular analysis of the gypsy chromatin insulator of Drosophila.Proc Natl Acad Sci U S A. 1996 Sep 3;93(18):9378-83. doi: 10.1073/pnas.93.18.9378. Proc Natl Acad Sci U S A. 1996. PMID: 8790337 Free PMC article. Review.
Cited by
-
The N-Terminal Part of Drosophila CP190 Is a Platform for Interaction with Multiple Architectural Proteins.Int J Mol Sci. 2023 Nov 2;24(21):15917. doi: 10.3390/ijms242115917. Int J Mol Sci. 2023. PMID: 37958900 Free PMC article.
-
New Drosophila promoter-associated architectural protein Mzfp1 interacts with CP190 and is required for housekeeping gene expression and insulator activity.Nucleic Acids Res. 2024 Jul 8;52(12):6886-6905. doi: 10.1093/nar/gkae393. Nucleic Acids Res. 2024. PMID: 38769058 Free PMC article.
-
Multiple Roles of dXNP and dADD1-Drosophila Orthologs of ATRX Chromatin Remodeler.Int J Mol Sci. 2023 Nov 18;24(22):16486. doi: 10.3390/ijms242216486. Int J Mol Sci. 2023. PMID: 38003676 Free PMC article. Review.
-
Chromatin insulator mechanisms ensure accurate gene expression by controlling overall 3D genome organization.Curr Opin Genet Dev. 2024 Aug;87:102208. doi: 10.1016/j.gde.2024.102208. Epub 2024 May 28. Curr Opin Genet Dev. 2024. PMID: 38810546 Free PMC article. Review.
-
Identification of a Highly Conserved Region Critical for the Functionality of the Cp190 Protein in Drosophila melanogaster.Dokl Biochem Biophys. 2025 Apr;521(1):178-182. doi: 10.1134/S1607672924601434. Epub 2025 Apr 11. Dokl Biochem Biophys. 2025. PMID: 40216711
References
-
- Jerkovic I., Szabo Q., Bantignies F., Cavalli G.. Higher-order chromosomal structures mediate genome function. J. Mol. Biol. 2020; 432:676–681. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

