Induction of senescence upon loss of the Ash2l core subunit of H3K4 methyltransferase complexes
- PMID: 35819198
- PMCID: PMC9371893
- DOI: 10.1093/nar/gkac591
Induction of senescence upon loss of the Ash2l core subunit of H3K4 methyltransferase complexes
Abstract
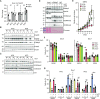
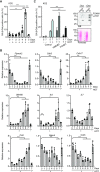
Gene expression is controlled in part by post-translational modifications of core histones. Methylation of lysine 4 of histone H3 (H3K4), associated with open chromatin and gene transcription, is catalyzed by type 2 lysine methyltransferase complexes that require WDR5, RBBP5, ASH2L and DPY30 as core subunits. Ash2l is essential during embryogenesis and for maintaining adult tissues. To expand on the mechanistic understanding of Ash2l, we generated mouse embryo fibroblasts (MEFs) with conditional Ash2l alleles. Upon loss of Ash2l, methylation of H3K4 and gene expression were downregulated, which correlated with inhibition of proliferation and cell cycle progression. Moreover, we observed induction of senescence concomitant with a set of downregulated signature genes but independent of SASP. Many of the signature genes are FoxM1 responsive. Indeed, exogenous FOXM1 was sufficient to delay senescence. Thus, although the loss of Ash2l in MEFs has broad and complex consequences, a distinct set of downregulated genes promotes senescence.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
Loss of the Ash2l subunit of histone H3K4 methyltransferase complexes reduces chromatin accessibility at promoters.Sci Rep. 2022 Dec 13;12(1):21506. doi: 10.1038/s41598-022-25881-0. Sci Rep. 2022. PMID: 36513698 Free PMC article.
-
An Ash2L/RbBP5 heterodimer stimulates the MLL1 methyltransferase activity through coordinated substrate interactions with the MLL1 SET domain.PLoS One. 2010 Nov 23;5(11):e14102. doi: 10.1371/journal.pone.0014102. PLoS One. 2010. PMID: 21124902 Free PMC article.
-
Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes.Nat Struct Mol Biol. 2006 Sep;13(9):852-4. doi: 10.1038/nsmb1131. Epub 2006 Aug 6. Nat Struct Mol Biol. 2006. PMID: 16892064
-
Diverse roles of WDR5-RbBP5-ASH2L-DPY30 (WRAD) complex in the functions of the SET1 histone methyltransferase family.J Biosci. 2017 Mar;42(1):155-159. doi: 10.1007/s12038-017-9666-9. J Biosci. 2017. PMID: 28229975 Review.
-
Modes of Interaction of KMT2 Histone H3 Lysine 4 Methyltransferase/COMPASS Complexes with Chromatin.Cells. 2018 Mar 2;7(3):17. doi: 10.3390/cells7030017. Cells. 2018. PMID: 29498679 Free PMC article. Review.
Cited by
-
Interrogating the regulatory epigenome of cellular senescence.Cell Mol Life Sci. 2025 Aug 31;82(1):328. doi: 10.1007/s00018-025-05848-w. Cell Mol Life Sci. 2025. PMID: 40886198 Free PMC article. Review.
-
Loss of the Ash2l subunit of histone H3K4 methyltransferase complexes reduces chromatin accessibility at promoters.Sci Rep. 2022 Dec 13;12(1):21506. doi: 10.1038/s41598-022-25881-0. Sci Rep. 2022. PMID: 36513698 Free PMC article.
-
Sequential deregulation of histone marks, chromatin accessibility and gene expression in response to PROTAC-induced degradation of ASH2L.Sci Rep. 2023 Dec 19;13(1):22565. doi: 10.1038/s41598-023-49284-x. Sci Rep. 2023. PMID: 38114530 Free PMC article.
-
Single-cell transcriptomics implicates the FEZ1-DKK1 axis in the regulation of corneal epithelial cell proliferation and senescence.Cell Prolif. 2023 Sep;56(9):e13433. doi: 10.1111/cpr.13433. Epub 2023 Feb 27. Cell Prolif. 2023. PMID: 36851859 Free PMC article.
-
KDM4C-mediated senescence defense is a targetable vulnerability in gastric cancer harboring TP53 mutations.Clin Epigenetics. 2023 Oct 17;15(1):163. doi: 10.1186/s13148-023-01579-6. Clin Epigenetics. 2023. PMID: 37848946 Free PMC article.
References
-
- Kornberg R.D., Lorch Y. Perspective primary role of the nucleosome. Mol. Cell. 2020; 79:371–375. - PubMed
-
- Zentner G.E., Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nat. Struct. Mol. Biol. 2013; 20:259–266. - PubMed
-
- Morgan M.A.J., Shilatifard A. Reevaluating the roles of histone-modifying enzymes and their associated chromatin modifications in transcriptional regulation. Nat. Genet. 2020; 52:1271–1281. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

