AACR Project GENIE: 100,000 Cases and Beyond
- PMID: 35819403
- PMCID: PMC9437568
- DOI: 10.1158/2159-8290.CD-21-1547
AACR Project GENIE: 100,000 Cases and Beyond
Abstract
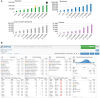
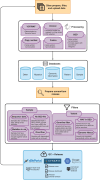
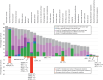
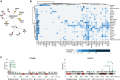
The American Association for Cancer Research (AACR) Project Genomics Evidence Neoplasia Information Exchange (GENIE) is an international pan-cancer registry with the goal to inform cancer research and clinical care worldwide. Founded in late 2015, the milestone GENIE 9.1-public release contains data from >110,000 tumors from >100,000 people treated at 19 cancer centers from the United States, Canada, the United Kingdom, France, the Netherlands, and Spain. Here, we demonstrate the use of these real-world data, harmonized through a centralized data resource, to accurately predict enrollment on genome-guided trials, discover driver alterations in rare tumors, and identify cancer types without actionable mutations that could benefit from comprehensive genomic analysis. The extensible data infrastructure and governance framework support additional deep patient phenotyping through biopharmaceutical collaborations and expansion to include new data types such as cell-free DNA sequencing. AACR Project GENIE continues to serve a global precision medicine knowledge base of increasing impact to inform clinical decision-making and bring together cancer researchers internationally.
Significance: AACR Project GENIE has now accrued data from >110,000 tumors, placing it among the largest repository of publicly available, clinically annotated genomic data in the world. GENIE has emerged as a powerful resource to evaluate genome-guided clinical trial design, uncover drivers of cancer subtypes, and inform real-world use of genomic data. This article is highlighted in the In This Issue feature, p. 2007.
©2022 The Authors; Published by the American Association for Cancer Research.
Figures


Comment in
References
-
- LeNoue-Newton ML, Chen SC, Stricker T, Hyman DM, Blauvelt N, Bedard PL, et al. Natural history and characteristics of ERBB2-mutated hormone receptor–positive metastatic breast cancer: a multi-institutional retrospective case–control study from AACR project GENIE. Clin Cancer Res 2022;28:2118–30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

