HumanMine: advanced data searching, analysis and cross-species comparison
- PMID: 35820040
- PMCID: PMC9275753
- DOI: 10.1093/database/baac054
HumanMine: advanced data searching, analysis and cross-species comparison
Abstract
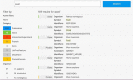
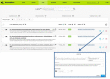
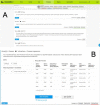
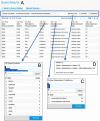
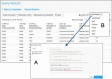
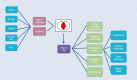
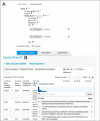
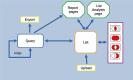
HumanMine (www.humanmine.org) is an integrated database of human genomics and proteomics data that provides a powerful interface to support sophisticated exploration and analysis of data compiled from experimental, computational and curated data sources. Built using the InterMine data integration platform, HumanMine includes genes, proteins, pathways, expression levels, Single nucleotide polymorphism (SNP), diseases and more, integrated into a single searchable database. HumanMine promotes integrative analysis, a powerful approach in modern biology that allows many sources of evidence to be analysed together. The data can be accessed through a user-friendly web interface as well as a powerful, scriptable web service Application programming interface (API) to allow programmatic access to data. The web interface includes a useful identifier resolution system, sophisticated query options and interactive results tables that enable powerful exploration of data, including data summaries, filtering, browsing and export. A set of graphical analysis tools provide a rich environment for data exploration including statistical enrichment of sets of genes or other biological entities. HumanMine can be used for integrative multistaged analysis that can lead to new insights and uncover previously unknown relationships. Database URL: https://www.humanmine.org.
© The Author(s) 2022. Published by Oxford University Press.
Figures





References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

