PU.1-Dependent Enhancer Inhibition Separates Tet2-Deficient Hematopoiesis from Malignant Transformation
- PMID: 35820129
- PMCID: PMC9894728
- DOI: 10.1158/2643-3230.BCD-21-0226
PU.1-Dependent Enhancer Inhibition Separates Tet2-Deficient Hematopoiesis from Malignant Transformation
Abstract
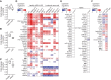
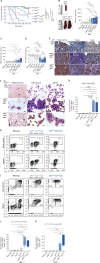
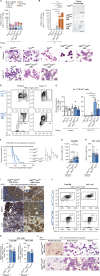
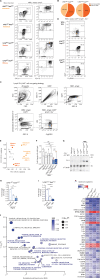
Cytosine hypermethylation in and around DNA-binding sites of master transcription factors, including PU.1, occurs in aging hematopoietic stem cells following acquired loss-of-function mutations of DNA methyl-cytosine dioxygenase ten-eleven translocation-2 (TET2), albeit functional relevance has been unclear. We show that Tet2-deficient mouse hematopoietic stem and progenitor cells undergo malignant transformation upon compromised gene regulation through heterozygous deletion of an upstream regulatory region (UREΔ/WT) of the PU.1 gene. Although compatible with multilineage blood formation at young age, Tet2-deficient PU.1 UREΔ/WT mice develop highly penetrant, transplantable acute myeloid leukemia (AML) during aging. Leukemic stem and progenitor cells show hypermethylation at putative PU.1-binding sites, fail to activate myeloid enhancers, and are hallmarked by a signature of genes with impaired expression shared with human AML. Our study demonstrates that Tet2 and PU.1 jointly suppress leukemogenesis and uncovers a methylation-sensitive PU.1-dependent gene network as a unifying molecular vulnerability associated with AML.
Significance: We identify moderately impaired PU.1 mRNA expression as a biological modality predisposing Tet2-deficient hematopoietic stem and progenitor cells to malignant transformation. Our study furthermore uncovers a methylation-sensitive PU.1 gene network as a common feature of myeloid leukemia potentially allowing for the identification of patients at risk for malignant transformation. See related commentary by Schleicher and Pietras, p. 378. This article is highlighted in the In This Issue feature, p. 369.
©2022 The Authors; Published by the American Association for Cancer Research.
Figures


Comment in
-
Reduced PU.1 Expression Collaborates with Tet2 Loss to Trigger Myeloid Leukemogenesis.Blood Cancer Discov. 2022 Sep 6;3(5):378-381. doi: 10.1158/2643-3230.BCD-22-0100. Blood Cancer Discov. 2022. PMID: 36065607 Free PMC article.
References
-
- Bonnefond A, Skrobek B, Lobbens S, Eury E, Thuillier D, Cauchi S, et al. Association between large detectable clonal mosaicism and type 2 diabetes with vascular complications. Nat Genet 2013;45:1040–3. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

