The role of gut microbial β-glucuronidase in drug disposition and development
- PMID: 35820618
- PMCID: PMC9717552
- DOI: 10.1016/j.drudis.2022.07.001
The role of gut microbial β-glucuronidase in drug disposition and development
Abstract
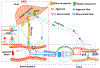
Gut microbial β-glucuronidase (gmGUS) is involved in the disposition of many endogenous and exogenous compounds. Preclinical studies have shown that inhibiting gmGUS activity affects drug disposition, resulting in reduced toxicity in the gastrointestinal tract (GIT) and enhanced systemic efficacy. Additionally, manipulating gmGUS activity is expected to be effective in preventing/treating local or systemic diseases. Although results from animal studies are promising, challenges remain in developing drugs by targeting gmGUS. Here, we review the role of gmGUS in host health under physiological and pathological conditions, the impact of gmGUS on the disposition of phenolic compounds, models used to study gmGUS activity, and the perspectives and challenges in developing drugs by targeting gmGUS.
Keywords: Drug development; Drug disposition; Druggable target; Gut microbial β-glucuronidase.
Copyright © 2022 Elsevier Ltd. All rights reserved.
Figures


Similar articles
-
Gut microbial beta-glucuronidase: a vital regulator in female estrogen metabolism.Gut Microbes. 2023 Jan-Dec;15(1):2236749. doi: 10.1080/19490976.2023.2236749. Gut Microbes. 2023. PMID: 37559394 Free PMC article. Review.
-
Gut microbial β-Glucuronidase: A key regulator of endobiotic homeostasis.Cell Host Microbe. 2024 Jun 12;32(6):783-785. doi: 10.1016/j.chom.2024.05.007. Cell Host Microbe. 2024. PMID: 38870895
-
Hinokiflavone from Platycladi cacumen as a potent broad-spectrum inhibitor of gut microbial Loop-1 β-glucuronidases: Inhibition kinetics and molecular simulation.Chem Biol Interact. 2024 Dec 1;404:111261. doi: 10.1016/j.cbi.2024.111261. Epub 2024 Oct 9. Chem Biol Interact. 2024. PMID: 39389440
-
Human gut bacterial β-glucuronidase inhibition: An emerging approach to manage medication therapy.Biochem Pharmacol. 2021 Aug;190:114566. doi: 10.1016/j.bcp.2021.114566. Epub 2021 Apr 16. Biochem Pharmacol. 2021. PMID: 33865833 Review.
-
β-Glucuronidase Inhibition in Drug Development: Emerging Strategies for Mitigating Drug-Induced Toxicity and Enhancing Therapeutic Outcomes.Drug Dev Res. 2025 Jun;86(4):e70118. doi: 10.1002/ddr.70118. Drug Dev Res. 2025. PMID: 40522235 Review.
Cited by
-
Microbiome and pancreatic cancer: time to think about chemotherapy.Gut Microbes. 2024 Jan-Dec;16(1):2374596. doi: 10.1080/19490976.2024.2374596. Epub 2024 Jul 18. Gut Microbes. 2024. PMID: 39024520 Free PMC article. Review.
-
Sex differences in opioid response: a role for the gut microbiome?Front Pharmacol. 2024 Aug 29;15:1455416. doi: 10.3389/fphar.2024.1455416. eCollection 2024. Front Pharmacol. 2024. PMID: 39268474 Free PMC article. Review.
-
Pinto Bean Supplementation Modulates Gut Microbiota and Improves Markers of Gut Integrity in a Mouse Model of Estrogen Deficiency.J Nutr. 2025 Jul 17:S0022-3166(25)00431-6. doi: 10.1016/j.tjnut.2025.07.008. Online ahead of print. J Nutr. 2025. PMID: 40680856 Free PMC article.
-
Discovering microbiota functions via chemical probe incorporation for targeted sequencing.Curr Opin Chem Biol. 2025 Feb;84:102551. doi: 10.1016/j.cbpa.2024.102551. Epub 2024 Nov 30. Curr Opin Chem Biol. 2025. PMID: 39615426 Review.
-
Dietary fibre optimisation in support of global health.Microb Biotechnol. 2024 Aug;17(8):e14542. doi: 10.1111/1751-7915.14542. Microb Biotechnol. 2024. PMID: 39096198 Free PMC article. Review.
References
-
- Winter J, Bokkenheuser VD. Bacterial metabolism of natural and synthetic sex hormones undergoing enterohepatic circulation. J Steroid Biochem; 27 (1987) 1145–1149. - PubMed
-
- Masamune H BIOCHEMICAL STUDIES ON CARBOHYDRATES: IV. On an Enzyme which Catalyses the Hydrolysis of Biosynthetic Osides of Glucuronic Acid. J Biochem; 19 (1934) 353–375.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

