Antagonistic effects of mitochondrial matrix and intermembrane space proteases on yeast aging
- PMID: 35820914
- PMCID: PMC9277893
- DOI: 10.1186/s12915-022-01352-w
Antagonistic effects of mitochondrial matrix and intermembrane space proteases on yeast aging
Abstract
Background: In many organisms, aging is characterized by a loss of mitochondrial homeostasis. Multiple factors such as respiratory metabolism, mitochondrial fusion/fission, or mitophagy have been linked to cell longevity, but the exact impact of each one on the aging process is still unclear.
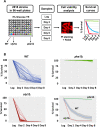
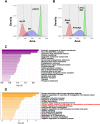
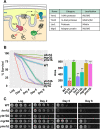
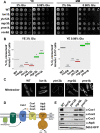
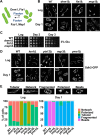
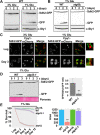
Results: Using the deletion mutant collection of the fission yeast Schizosaccharomyces pombe, we have developed a genome-wide screening for mutants with altered chronological lifespan. We have identified four mutants associated with proteolysis at the mitochondria that exhibit opposite effects on longevity. The analysis of the respiratory activity of these mutants revealed a positive correlation between increased respiration rate and prolonged lifespan. We also found that the phenotype of the long-lived protease mutants could not be explained by impaired mitochondrial fusion/fission activities, but it was dependent on mitophagy induction. The anti-aging role of mitophagy was supported by the effect of a mutant defective in degradation of mitochondria, which shortened lifespan of the long-lived mutants.
Conclusions: Our characterization of the mitochondrial protease mutants demonstrates that mitophagy sustains the lifespan extension of long-lived mutants displaying a higher respiration potential.
Keywords: Chronological aging; Mitochondrial dynamics; Mitophagy; Mitoproteases; Respiratory capacity.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Genetic approaches to aging in budding and fission yeasts: new connections and new opportunities.Subcell Biochem. 2012;57:291-314. doi: 10.1007/978-94-007-2561-4_13. Subcell Biochem. 2012. PMID: 22094427 Review.
-
Simultaneous impairment of mitochondrial fission and fusion reduces mitophagy and shortens replicative lifespan.Sci Rep. 2015 Jan 20;5:7885. doi: 10.1038/srep07885. Sci Rep. 2015. PMID: 25601284 Free PMC article.
-
Inhibition of peroxisome fission, but not mitochondrial fission, increases yeast chronological lifespan.Cell Cycle. 2015;14(11):1698-703. doi: 10.1080/15384101.2015.1029685. Cell Cycle. 2015. PMID: 25840089 Free PMC article.
-
Mitophagy in yeast: Molecular mechanisms and physiological role.Biochim Biophys Acta. 2015 Oct;1853(10 Pt B):2756-65. doi: 10.1016/j.bbamcr.2015.01.005. Epub 2015 Jan 17. Biochim Biophys Acta. 2015. PMID: 25603537 Review.
-
Dual roles of mitochondrial fusion gene FZO1 in yeast age asymmetry and in longevity mediated by a novel ATG32-dependent retrograde response.Biogerontology. 2019 Feb;20(1):93-107. doi: 10.1007/s10522-018-9779-z. Epub 2018 Oct 8. Biogerontology. 2019. PMID: 30298458
Cited by
-
Imaging of mtHyPer7, a Ratiometric Biosensor for Mitochondrial Peroxide, in Living Yeast Cells.J Vis Exp. 2023 Jun 2;(196):10.3791/65428. doi: 10.3791/65428. J Vis Exp. 2023. PMID: 37335116 Free PMC article.
-
Novel TORC1 inhibitor Ecl1 is regulated by phosphorylation in fission yeast.Aging Cell. 2025 Apr;24(4):e14450. doi: 10.1111/acel.14450. Epub 2025 Feb 5. Aging Cell. 2025. PMID: 39910760 Free PMC article.
-
Comparing Mitochondrial Activity, Oxidative Stress Tolerance, and Longevity of Thirteen Ascomycota Yeast Species.Antioxidants (Basel). 2023 Sep 28;12(10):1810. doi: 10.3390/antiox12101810. Antioxidants (Basel). 2023. PMID: 37891889 Free PMC article.
-
Mitochondrial proteases and their roles in mitophagy in plants, animals, and yeast.Plant Cell Physiol. 2025 Aug 12;66(7):973-990. doi: 10.1093/pcp/pcaf038. Plant Cell Physiol. 2025. PMID: 40269524 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

