Whole-genome sequence analysis reveals selection signatures for important economic traits in Xiang pigs
- PMID: 35821031
- PMCID: PMC9276726
- DOI: 10.1038/s41598-022-14686-w
Whole-genome sequence analysis reveals selection signatures for important economic traits in Xiang pigs
Abstract
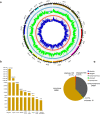
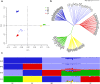
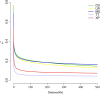
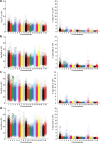
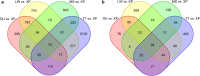
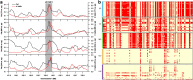
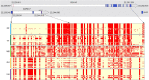
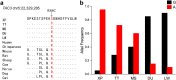
Xiang pig (XP) is one of the best-known indigenous pig breeds in China, which is characterized by its small body size, strong disease resistance, high adaptability, favorite meat quality, small litter sizes, and early sexual maturity. However, the genomic evidence that links these unique traits of XP is still poorly understood. To identify the genomic signatures of selection in XP, we performed whole-genome resequencing on 25 unrelated individual XPs. We obtained 876.70 Gb of raw data from the genomic libraries. The LD analysis showed that the lowest level of linkage disequilibrium was observed in Xiang pig. Comparative genomic analysis between XPs and other breeds including Tibetan, Meishan, Duroc and Landrace revealed 3062, 1228, 907 and 1519 selected regions, respectively. The genes identified in selected regions of XPs were associated with growth and development processes (IGF1R, PROP1, TBX19, STAC3, RLF, SELENOM, MSTN), immunity and disease resistance (ZCCHC2, SERPINB2, ADGRE5, CYP7B1, STAT6, IL2, CD80, RHBDD3, PIK3IP1), environmental adaptation (NR2E1, SERPINB8, SERPINB10, SLC26A7, MYO1A, SDR9C7, UVSSA, EXPH5, VEGFC, PDE1A), reproduction (CCNB2, TRPM6, EYA3, CYP7B1, LIMK2, RSPO1, ADAM32, SPAG16), meat quality traits (DECR1, EWSR1), and early sexual maturity (TAC3). Through the absolute allele frequency difference (ΔAF) analysis, we explored two population-specific missense mutations occurred in NR6A1 and LTBP2 genes, which well explained that the vertebrae numbers of Xiang pigs were less than that of the European pig breeds. Our results indicated that Xiang pigs were less affected by artificial selection than the European and Meishan pig breeds. The selected candidate genes were mainly involved in growth and development, disease resistance, reproduction, meat quality, and early sexual maturity. This study provided a list of functional candidate genes, as well as a number of genetic variants, which would provide insight into the molecular basis for the unique traits of Xiang pig.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Identification of Copy Number Variations in Xiang and Kele Pigs.PLoS One. 2016 Feb 3;11(2):e0148565. doi: 10.1371/journal.pone.0148565. eCollection 2016. PLoS One. 2016. PMID: 26840413 Free PMC article.
-
The selected genes NR6A1, RSAD2-CMPK2, and COL3A1 contribute to body size variation in Meishan pigs through different patterns.J Anim Sci. 2023 Jan 3;101:skad304. doi: 10.1093/jas/skad304. J Anim Sci. 2023. PMID: 37703114 Free PMC article.
-
Whole-genome sequencing of European autochthonous and commercial pig breeds allows the detection of signatures of selection for adaptation of genetic resources to different breeding and production systems.Genet Sel Evol. 2020 Jun 26;52(1):33. doi: 10.1186/s12711-020-00553-7. Genet Sel Evol. 2020. PMID: 32591011 Free PMC article.
-
Assessing genomic diversity and signatures of selection in Jiaxian Red cattle using whole-genome sequencing data.BMC Genomics. 2021 Jan 9;22(1):43. doi: 10.1186/s12864-020-07340-0. BMC Genomics. 2021. PMID: 33421990 Free PMC article. Review.
-
Genome-wide selection signatures detection in Shanghai Holstein cattle population identified genes related to adaption, health and reproduction traits.BMC Genomics. 2021 Oct 15;22(1):747. doi: 10.1186/s12864-021-08042-x. BMC Genomics. 2021. PMID: 34654366 Free PMC article. Review.
Cited by
-
Identification of candidate genomic regions for egg yolk moisture content based on a genome-wide association study.BMC Genomics. 2023 Mar 14;24(1):110. doi: 10.1186/s12864-023-09221-8. BMC Genomics. 2023. PMID: 36918797 Free PMC article.
-
Pangenome Reveals Gene Content Variations and Structural Variants Contributing to Pig Characteristics.Genomics Proteomics Bioinformatics. 2025 Jan 15;22(6):qzae081. doi: 10.1093/gpbjnl/qzae081. Genomics Proteomics Bioinformatics. 2025. PMID: 39535885 Free PMC article.
-
Applications of Next-Generation Sequencing Technologies and Statistical Tools in Identifying Pathways and Biomarkers for Heat Tolerance in Livestock.Vet Sci. 2024 Dec 2;11(12):616. doi: 10.3390/vetsci11120616. Vet Sci. 2024. PMID: 39728955 Free PMC article. Review.
-
Genome-Wide Association Study Meta-Analysis Elucidates Genetic Structure and Identifies Candidate Genes of Teat Number Traits in Pigs.Int J Mol Sci. 2023 Dec 29;25(1):451. doi: 10.3390/ijms25010451. Int J Mol Sci. 2023. PMID: 38203622 Free PMC article.
-
Integrative Analysis of Transcriptomic and Lipidomic Profiles Reveals a Differential Subcutaneous Adipose Tissue Mechanism among Ningxiang Pig and Berkshires, and Their Offspring.Animals (Basel). 2023 Oct 25;13(21):3321. doi: 10.3390/ani13213321. Animals (Basel). 2023. PMID: 37958077 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

