Multiplex Hextuple Luciferase Assaying
- PMID: 35821491
- PMCID: PMC10157609
- DOI: 10.1007/978-1-0716-2453-1_33
Multiplex Hextuple Luciferase Assaying
Abstract
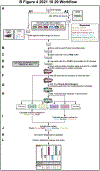
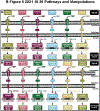
We recently expanded the commonly used dual luciferase assaying method toward multiplex hextuple luciferase assaying, allowing monitoring the activity of five experimental pathways against one control at the same time. In doing so, while our expanded assay utilizes a total of six orthogonal luciferases instead of two, this assay, conveniently, still utilizes the well-established reagents and principles of the widely used dual luciferase assay. Three quenchable D-luciferin-consuming luciferases are measured after addition of D-Luciferin substrate, followed by quenching of their bioluminescence (BL) and the measurement of three coelenterazine (CTZ)-consuming luciferases after addition of CTZ substrate, all in the same vessel. Here, we provide detailed protocols on how to perform such multiplex hextuple luciferase assaying to monitor cellular signal processing upstream of five transcription factors and their corresponding transcription factor-binding motifs, using a constitutive promoter as normalization control. The first protocol is provided on how to perform cell culture in preparation toward genetic or pharmaceutical perturbations, as well as transfecting a multiplex hextuple luciferase reporter vector encoding all luciferase reporter units needed for multiplex hextuple luciferase assaying. The second protocol details on how to execute multiplex hextuple luciferase assaying using a microplate reader appropriately equipped to detect the different BLs emitted by all six luciferases. Finally, the third protocol provides details on analyzing, plotting, and interpreting the data obtained by the microplate reader.
Keywords: Assay; Cell culture; Cellular signaling pathway; Hextuple; Luciferase; Microplate reader; Multiplex; Orthogonal; Pathway perturbation; Transfection.
© 2022. The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature.
Figures











References
-
- Berg EL (2017) Phenotypic chemical biology for predicting safety and efficacy. Drug Discov Today Technol 23:53–60 - PubMed
-
- Taylor DL and Giuliano KA (2005) Multiplexed high content screening assays create a systems cell biology approach to drug discovery. Drug Discov Today Suppl:13–18 - PubMed
-
- Westwick JK and Lamerdin JE (2011) Improving drug discovery with contextual assays and cellular systems analysis. Methods Mol Biol 756:61–73 - PubMed
-
- Korn K and Krausz E (2007) Cell-based high-content screening of small-molecule libraries. Curr Opin Chem Biol 11:503–510 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

