Adapting Blood DNA Methylation Aging Clocks for Use in Saliva Samples With Cell-type Deconvolution
- PMID: 35822029
- PMCID: PMC9261380
- DOI: 10.3389/fragi.2021.697254
Adapting Blood DNA Methylation Aging Clocks for Use in Saliva Samples With Cell-type Deconvolution
Abstract
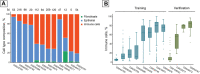
DeepMAge is a deep-learning DNA methylation aging clock that measures the organismal pace of aging with the information from human epigenetic profiles. In blood samples, DeepMAge can predict chronological age within a 2.8 years error margin, but in saliva samples, its performance is drastically reduced since aging clocks are restricted by the training set domain. However, saliva is an attractive fluid for genomic studies due to its availability, compared to other tissues, including blood. In this article, we display how cell type deconvolution and elastic net can be used to expand the domain of deep aging clocks to other tissues. Using our approach, DeepMAge's error in saliva samples was reduced from 20.9 to 4.7 years with no retraining.
Keywords: DNA methylation; aging; aging clock; biogerontology; cell-type deconvolution; deep learning; domain adaptation; epigenetics.
Copyright © 2021 Galkin, Kochetov, Mamoshina and Zhavoronkov.
Conflict of interest statement
FG, KK, AZ, PM were employed by Deep Longevity Limited - part of Endurance RP Limited (SEHK: 0575.HK). AZ was employed by Insilico Medicine Hong Kong Limited. DeepMAge is a patent-pending aging clock.
Figures



References
LinkOut - more resources
Full Text Sources

