Evolutionary traits of Tick-borne encephalitis virus: Pervasive non-coding RNA structure conservation and molecular epidemiology
- PMID: 35822110
- PMCID: PMC9272599
- DOI: 10.1093/ve/veac051
Evolutionary traits of Tick-borne encephalitis virus: Pervasive non-coding RNA structure conservation and molecular epidemiology
Abstract
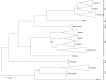
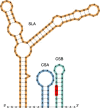
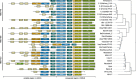
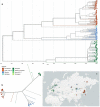
Tick-borne encephalitis virus (TBEV) is the aetiological agent of tick-borne encephalitis, an infectious disease of the central nervous system that is often associated with severe sequelae in humans. While TBEV is typically classified into three subtypes, recent evidence suggests a more varied range of TBEV subtypes and lineages that differ substantially in the architecture of their 3' untranslated region (3'UTR). Building on comparative genomic approaches and thermodynamic modelling, we characterize the TBEV UTR structureome diversity and propose a unified picture of pervasive non-coding RNA structure conservation. Moreover, we provide an updated phylogeny of TBEV, building on more than 220 publicly available complete genomes, and investigate the molecular epidemiology and phylodynamics with Nextstrain, a web-based visualization framework for real-time pathogen evolution.
Keywords: Tick-borne encephalitis virus; conserved RNA structure; molecular epidemiology; untranslated region.
© The Author(s) 2022. Published by Oxford University Press.
Figures
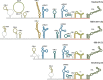
References
-
- Adelshin R. V. et al. (2019) ‘‘886-84-Like’ Tick-Borne Encephalitis Virus Strains: Intraspecific Status Elucidated by Comparative Genomics’, Ticks and Tick-borne Diseases, 10: 1168–72. - PubMed
LinkOut - more resources
Full Text Sources

