A systematic evaluation of the computational tools for ligand-receptor-based cell-cell interaction inference
- PMID: 35822343
- PMCID: PMC9479691
- DOI: 10.1093/bfgp/elac019
A systematic evaluation of the computational tools for ligand-receptor-based cell-cell interaction inference
Abstract
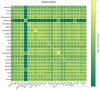
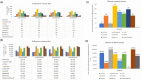
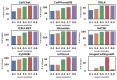
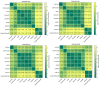
Cell-cell interactions (CCIs) are essential for multicellular organisms to coordinate biological processes and functions. One classical type of CCI interaction is between secreted ligands and cell surface receptors, i.e. ligand-receptor (LR) interactions. With the recent development of single-cell technologies, a large amount of single-cell ribonucleic acid (RNA) sequencing (scRNA-Seq) data has become widely available. This data availability motivated the single-cell-resolution study of CCIs, particularly LR-based CCIs. Dozens of computational methods and tools have been developed to predict CCIs by identifying LR-based CCIs. Many of these tools have been theoretically reviewed. However, there is little study on current LR-based CCI prediction tools regarding their performance and running results on public scRNA-Seq datasets. In this work, to fill this gap, we tested and compared nine of the most recent computational tools for LR-based CCI prediction. We used 15 well-studied scRNA-Seq samples that correspond to approximately 100K single cells under different experimental conditions for testing and comparison. Besides briefing the methodology used in these nine tools, we summarized the similarities and differences of these tools in terms of both LR prediction and CCI inference between cell types. We provided insight into using these tools to make meaningful discoveries in understanding cell communications.
Keywords: cell–cell interaction; computational prediction tools; ligand-receptor interaction; single-cell RNA sequencing.
© The Author(s) 2022. Published by Oxford University Press.
Figures

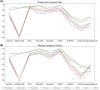

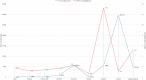
References
-
- Efremova M, Vento-Tormo M, Teichmann SA, et al. . CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc 2020;15:1484–506. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

