Using adopted individuals to partition indirect maternal genetic effects into prenatal and postnatal effects on offspring phenotypes
- PMID: 35822614
- PMCID: PMC9323003
- DOI: 10.7554/eLife.73671
Using adopted individuals to partition indirect maternal genetic effects into prenatal and postnatal effects on offspring phenotypes
Abstract
Maternal genetic effects can be defined as the effect of a mother's genotype on the phenotype of her offspring, independent of the offspring's genotype. Maternal genetic effects can act via the intrauterine environment during pregnancy and/or via the postnatal environment. In this manuscript, we present a simple extension to the basic adoption design that uses structural equation modelling (SEM) to partition maternal genetic effects into prenatal and postnatal effects. We examine the power, utility and type I error rate of our model using simulations and asymptotic power calculations. We apply our model to polygenic scores of educational attainment and birth weight associated variants, in up to 5,178 adopted singletons, 943 trios, 2687 mother-offspring pairs, 712 father-offspring pairs and 347,980 singletons from the UK Biobank. Our results show the expected pattern of maternal genetic effects on offspring birth weight, but unexpectedly large prenatal maternal genetic effects on offspring educational attainment. Sensitivity and simulation analyses suggest this result may be at least partially due to adopted individuals in the UK Biobank being raised by their biological relatives. We show that accurate modelling of these sorts of cryptic relationships is sufficient to bring type I error rate under control and produce asymptotically unbiased estimates of prenatal and postnatal maternal genetic effects. We conclude that there would be considerable value in following up adopted individuals in the UK Biobank to determine whether they were raised by their biological relatives, and if so, to precisely ascertain the nature of these relationships. These adopted individuals could then be incorporated into informative statistical genetics models like the one described in our manuscript to further elucidate the genetic architecture of complex traits and diseases.
Keywords: adoption study; genetics; genomics; human; indirect maternal genetic effects; maternal genetic effects; postnatal maternal effects; prenatal maternal effects; structural equation modelling.
© 2022, Hwang et al.
Conflict of interest statement
LH, GM, DE No competing interests declared
Figures


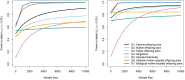



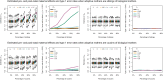








References
-
- Armstrong-Carter E, Trejo S, Hill LJB, Crossley KL, Mason D, Domingue BW. The earliest origins of genetic nurture the prenatal environment mediates the association between maternal genetics and child development. Psychological Science. 2020;31:781–791. doi: 10.1177/0956797620917209. - DOI - PMC - PubMed
-
- Bates TC, Maher BS, Medland SE, McAloney K, Wright MJ, Hansell NK, Kendler KS, Martin NG, Gillespie NA. The nature of nurture: using a virtual-parent design to test parenting effects on children’s educational attainment in genotyped families. Twin Research and Human Genetics. 2018;21:73–83. doi: 10.1017/thg.2018.11. - DOI - PubMed
-
- Beaumont RN, Warrington NM, Cavadino A, Tyrrell J, Nodzenski M, Horikoshi M, Geller F, Myhre R, Richmond RC, Paternoster L, Bradfield JP, Kreiner-Møller E, Huikari V, Metrustry S, Lunetta KL, Painter JN, Hottenga JJ, Allard C, Barton SJ, Espinosa A, Marsh JA, Potter C, Zhang G, Ang W, Berry DJ, Bouchard L, Das S, Hakonarson H, Heikkinen J, Helgeland Ø, Hocher B, Hofman A, Inskip HM, Jones SE, Kogevinas M, Lind PA, Marullo L, Medland SE, Murray A, Murray JC, Njølstad PR, Nohr EA, Reichetzeder C, Ring SM, Ruth KS, Santa-Marina L, Scholtens DM, Sebert S, Sengpiel V, Tuke MA, Vaudel M, Weedon MN, Willemsen G, Wood AR, Yaghootkar H, Muglia LJ, Bartels M, Relton CL, Pennell CE, Chatzi L, Estivill X, Holloway JW, Boomsma DI, Montgomery GW, Murabito JM, Spector TD, Power C, Järvelin MR, Bisgaard H, Grant SFA, Sørensen TIA, Jaddoe VW, Jacobsson B, Melbye M, McCarthy MI, Hattersley AT, Hayes MG, Frayling TM, Hivert MF, Felix JF, Hyppönen E, Lowe WL, Evans DM, Lawlor DA, Feenstra B, Freathy RM. Genome-wide association study of offspring birth weight in 86 577 women identifies five novel loci and highlights maternal genetic effects that are independent of fetal genetics. Human Molecular Genetics. 2018;27:742–756. doi: 10.1093/hmg/ddx429. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

