Enhanced polymerase activity permits efficient synthesis by cancer-associated DNA polymerase ϵ variants at low dNTP levels
- PMID: 35822874
- PMCID: PMC9371911
- DOI: 10.1093/nar/gkac602
Enhanced polymerase activity permits efficient synthesis by cancer-associated DNA polymerase ϵ variants at low dNTP levels
Abstract
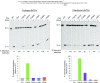
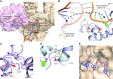
Amino acid substitutions in the exonuclease domain of DNA polymerase ϵ (Polϵ) cause ultramutated tumors. Studies in model organisms suggested pathogenic mechanisms distinct from a simple loss of exonuclease. These mechanisms remain unclear for most recurrent Polϵ mutations. Particularly, the highly prevalent V411L variant remained a long-standing puzzle with no detectable mutator effect in yeast despite the unequivocal association with ultramutation in cancers. Using purified four-subunit yeast Polϵ, we assessed the consequences of substitutions mimicking human V411L, S459F, F367S, L424V and D275V. While the effects on exonuclease activity vary widely, all common cancer-associated variants have increased DNA polymerase activity. Notably, the analog of Polϵ-V411L is among the strongest polymerases, and structural analysis suggests defective polymerase-to-exonuclease site switching. We further show that the V411L analog produces a robust mutator phenotype in strains that lack mismatch repair, indicating a high rate of replication errors. Lastly, unlike wild-type and exonuclease-dead Polϵ, hyperactive variants efficiently synthesize DNA at low dNTP concentrations. We propose that this characteristic could promote cancer cell survival and preferential participation of mutator polymerases in replication during metabolic stress. Our results support the notion that polymerase fitness, rather than low fidelity alone, is an important determinant of variant pathogenicity.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures







References
-
- Morrison A., Araki H., Clark A.B., Hamatake R.K., Sugino A.. A third essential DNA polymerase in .S. cerevisiae. Cell. 1990; 62:1143–1151. - PubMed
-
- Kunkel T.A. DNA replication fidelity. J. Biol. Chem. 2004; 279:16895–16898. - PubMed
-
- Ganai R.A., Johansson E.. DNA replication—a matter of fidelity. Mol. Cell. 2016; 62:745–755. - PubMed
-
- Pavlov Y.I., Zhuk A.S., Stepchenkova E.I.. DNA polymerases at the eukaryotic replication fork thirty years after: connection to cancer. Cancers (Basel). 2020; 12:3489.
-
- Reha-Krantz L.J. DNA polymerase proofreading: multiple roles maintain genome stability. Biochim. Biophys. Acta. 2010; 1804:1049–1063. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

