Cryo-EM structures reveal that RFC recognizes both the 3'- and 5'-DNA ends to load PCNA onto gaps for DNA repair
- PMID: 35829698
- PMCID: PMC9293004
- DOI: 10.7554/eLife.77469
Cryo-EM structures reveal that RFC recognizes both the 3'- and 5'-DNA ends to load PCNA onto gaps for DNA repair
Abstract
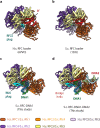
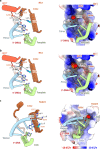
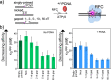
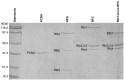
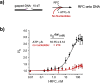
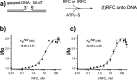
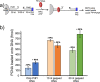
RFC uses ATP to assemble PCNA onto primed sites for replicative DNA polymerases δ and ε. The RFC pentamer forms a central chamber that binds 3' ss/ds DNA junctions to load PCNA onto DNA during replication. We show here five structures that identify a second DNA binding site in RFC that binds a 5' duplex. This 5' DNA site is located between the N-terminal BRCT domain and AAA+ module of the large Rfc1 subunit. Our structures reveal ideal binding to a 7-nt gap, which includes 2 bp unwound by the clamp loader. Biochemical studies show enhanced binding to 5 and 10 nt gaps, consistent with the structural results. Because both 3' and 5' ends are present at a ssDNA gap, we propose that the 5' site facilitates RFC's PCNA loading activity at a DNA damage-induced gap to recruit gap-filling polymerases. These findings are consistent with genetic studies showing that base excision repair of gaps greater than 1 base requires PCNA and involves the 5' DNA binding domain of Rfc1. We further observe that a 5' end facilitates PCNA loading at an RPA coated 30-nt gap, suggesting a potential role of the RFC 5'-DNA site in lagging strand DNA synthesis.
Keywords: DNA damage; DNA repair; DNA replication; PCNA; RFC; S. cerevisiae; biochemistry; chemical biology; sliding clamp.
© 2022, Zheng et al.
Conflict of interest statement
FZ, NY, HL, MO The authors declare no competing interests, RG No competing interests declared
Figures









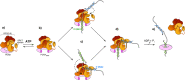
Similar articles
-
Cryo-EM reveals a nearly complete PCNA loading process and unique features of the human alternative clamp loader CTF18-RFC.Proc Natl Acad Sci U S A. 2024 Apr 30;121(18):e2319727121. doi: 10.1073/pnas.2319727121. Epub 2024 Apr 26. Proc Natl Acad Sci U S A. 2024. PMID: 38669181 Free PMC article.
-
Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.Elife. 2022 Feb 18;11:e74175. doi: 10.7554/eLife.74175. Elife. 2022. PMID: 35179493 Free PMC article.
-
Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification on biology.Cell Rep. 2023 Jul 25;42(7):112694. doi: 10.1016/j.celrep.2023.112694. Epub 2023 Jun 30. Cell Rep. 2023. PMID: 37392384 Free PMC article.
-
Functions of Multiple Clamp and Clamp-Loader Complexes in Eukaryotic DNA Replication.Adv Exp Med Biol. 2017;1042:135-162. doi: 10.1007/978-981-10-6955-0_7. Adv Exp Med Biol. 2017. PMID: 29357057 Review.
-
The PCNA-RFC families of DNA clamps and clamp loaders.Prog Nucleic Acid Res Mol Biol. 2004;78:227-60. doi: 10.1016/S0079-6603(04)78006-X. Prog Nucleic Acid Res Mol Biol. 2004. PMID: 15210332 Review.
Cited by
-
Differences in clamp loader mechanism between bacteria and eukaryotes.bioRxiv [Preprint]. 2023 Nov 30:2023.11.30.569468. doi: 10.1101/2023.11.30.569468. bioRxiv. 2023. Update in: J Biol Chem. 2024 Apr;300(4):107166. doi: 10.1016/j.jbc.2024.107166. PMID: 38076975 Free PMC article. Updated. Preprint.
-
Cryo-EM reveals a nearly complete PCNA loading process and unique features of the human alternative clamp loader CTF18-RFC.Proc Natl Acad Sci U S A. 2024 Apr 30;121(18):e2319727121. doi: 10.1073/pnas.2319727121. Epub 2024 Apr 26. Proc Natl Acad Sci U S A. 2024. PMID: 38669181 Free PMC article.
-
The human ATAD5 has evolved unique structural elements to function exclusively as a PCNA unloader.Nat Struct Mol Biol. 2024 Nov;31(11):1680-1691. doi: 10.1038/s41594-024-01332-4. Epub 2024 Jun 13. Nat Struct Mol Biol. 2024. PMID: 38871854 Free PMC article.
-
The proofreading mechanism of the human leading-strand DNA polymerase ε holoenzyme.Proc Natl Acad Sci U S A. 2025 Jun 3;122(22):e2507232122. doi: 10.1073/pnas.2507232122. Epub 2025 May 29. Proc Natl Acad Sci U S A. 2025. PMID: 40440070 Free PMC article.
-
From the Cytoplasm into the Nucleus-Hepatitis B Virus Travel and Genome Repair.Microorganisms. 2025 Jan 14;13(1):157. doi: 10.3390/microorganisms13010157. Microorganisms. 2025. PMID: 39858925 Free PMC article. Review.
References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica. Section D, Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

