What are housekeeping genes?
- PMID: 35830477
- PMCID: PMC9312424
- DOI: 10.1371/journal.pcbi.1010295
What are housekeeping genes?
Abstract
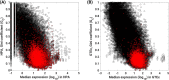
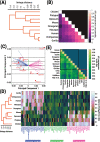
The concept of "housekeeping gene" has been used for four decades but remains loosely defined. Housekeeping genes are commonly described as "essential for cellular existence regardless of their specific function in the tissue or organism", and "stably expressed irrespective of tissue type, developmental stage, cell cycle state, or external signal". However, experimental support for the tenet that gene essentiality is linked to stable expression across cell types, conditions, and organisms has been limited. Here we use genome-scale functional genomic screens together with bulk and single-cell sequencing technologies to test this link and optimize a quantitative and experimentally validated definition of housekeeping gene. Using the optimized definition, we identify, characterize, and provide as resources, housekeeping gene lists extracted from several human datasets, and 10 other animal species that include primates, chicken, and C. elegans. We find that stably expressed genes are not necessarily essential, and that the individual genes that are essential and stably expressed can considerably differ across organisms; yet the pathways enriched among these genes are conserved. Further, the level of conservation of housekeeping genes across the analyzed organisms captures their taxonomic groups, showing evolutionary relevance for our definition. Therefore, we present a quantitative and experimentally supported definition of housekeeping genes that can contribute to better understanding of their unique biological and evolutionary characteristics.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Systematic Identification of Housekeeping Genes Possibly Used as References in Caenorhabditis elegans by Large-Scale Data Integration.Cells. 2020 Mar 24;9(3):786. doi: 10.3390/cells9030786. Cells. 2020. PMID: 32213971 Free PMC article.
-
Genome-wide identification of housekeeping genes in maize.Plant Mol Biol. 2014 Nov;86(4-5):543-54. doi: 10.1007/s11103-014-0246-1. Epub 2014 Sep 11. Plant Mol Biol. 2014. PMID: 25209110
-
Concept development of housekeeping genes in the high-throughput sequencing era.Yi Chuan. 2017 Feb 20;39(2):127-134. doi: 10.16288/j.yczz.16-379. Yi Chuan. 2017. PMID: 28242599 Review.
-
Machine learning approach to gene essentiality prediction: a review.Brief Bioinform. 2021 Sep 2;22(5):bbab128. doi: 10.1093/bib/bbab128. Brief Bioinform. 2021. PMID: 33842944 Review.
-
Comparing the evolutionary conservation between human essential genes, human orthologs of mouse essential genes and human housekeeping genes.Brief Bioinform. 2015 Nov;16(6):922-31. doi: 10.1093/bib/bbv025. Epub 2015 Apr 23. Brief Bioinform. 2015. PMID: 25911641
Cited by
-
Comparative Pan- and Phylo-Genomic Analysis of Ideonella and Thermobifida Strains: Dissemination of Biodegradation Potential and Genomic Divergence.Biochem Genet. 2025 Feb 25. doi: 10.1007/s10528-025-11031-4. Online ahead of print. Biochem Genet. 2025. PMID: 40000572
-
Refined culture conditions with increased physiological relevance uncover oncogene-dependent metabolic signatures in Ewing sarcoma spheroids.Cell Rep Methods. 2025 Feb 24;5(2):100966. doi: 10.1016/j.crmeth.2025.100966. Epub 2025 Feb 7. Cell Rep Methods. 2025. PMID: 39922188 Free PMC article.
-
Overexpression of housekeeping gene FveIPT2 enhances anthocyanin and terpenoid accumulation in strawberry fruits with minimal impact on plant growth and development.Hortic Res. 2025 May 26;12(8):uhaf130. doi: 10.1093/hr/uhaf130. eCollection 2025 Aug. Hortic Res. 2025. PMID: 40677761 Free PMC article.
-
Logical design of synthetic cis-regulatory DNA for genetic tracing of cell identities and state changes.Nat Commun. 2024 Feb 5;15(1):897. doi: 10.1038/s41467-024-45069-6. Nat Commun. 2024. PMID: 38316783 Free PMC article.
-
Transcriptional repression and enhancer decommissioning silence cell cycle genes in postmitotic tissues.G3 (Bethesda). 2024 Oct 7;14(10):jkae203. doi: 10.1093/g3journal/jkae203. G3 (Bethesda). 2024. PMID: 39171889 Free PMC article.

