Transition Path Sampling Based Calculations of Free Energies for Enzymatic Reactions: The Case of Human Methionine Adenosyl Transferase and Plasmodium vivax Adenosine Deaminase
- PMID: 35830574
- PMCID: PMC9444332
- DOI: 10.1021/acs.jpcb.2c03251
Transition Path Sampling Based Calculations of Free Energies for Enzymatic Reactions: The Case of Human Methionine Adenosyl Transferase and Plasmodium vivax Adenosine Deaminase
Abstract
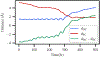
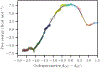
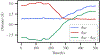
Transition path sampling (TPS) is widely used for the calculations of reaction rates, transition state structures, and reaction coordinates of condensed phase systems. Here we discuss a scheme for the calculation of free energies using the ensemble of TPS reactive trajectories in combination with a window-based sampling technique for enzyme-catalyzed reactions. We calculate the free energy profiles of the reactions catalyzed by the human methionine S-adenosyltransferase (MAT2A) enzyme and the Plasmodium vivax adenosine deaminase (pvADA) enzyme to assess the accuracy of this method. MAT2A catalyzes the formation of S-adenosine-l-methionine following a SN2 mechanism, and using our method, we estimate the free energy barrier for this reaction to be 16 kcal mol-1, which is in excellent agreement with the experimentally measured activation energy of 17.27 kcal mol-1. Furthermore, for the pvADA enzyme-catalyzed reaction we estimate a free energy barrier of 21 kcal mol-1, and the calculated free energy profile is similar to that predicted from experimental observations. Calculating free energies by employing our simple method within TPS provides significant advantages over methods such as umbrella sampling because it is free from any applied external bias, is accurate compared to experimental measurements, and has a reasonable computational cost.
Figures
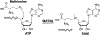




Similar articles
-
The Transition-State Structure for Human MAT2A from Isotope Effects.J Am Chem Soc. 2017 Oct 4;139(39):13754-13760. doi: 10.1021/jacs.7b05803. Epub 2017 Sep 20. J Am Chem Soc. 2017. PMID: 28880543 Free PMC article.
-
What make malarial adenosine deaminase from PLASMODIUM VIVAX recognise adenosine and 5'-methylthioadenosine: simulation studies.J Biomol Struct Dyn. 2023 Mar;41(4):1437-1444. doi: 10.1080/07391102.2021.2021989. Epub 2022 Jan 7. J Biomol Struct Dyn. 2023. PMID: 34994283
-
Quantum mechanics/molecular mechanics minimum free-energy path for accurate reaction energetics in solution and enzymes: sequential sampling and optimization on the potential of mean force surface.J Chem Phys. 2008 Jan 21;128(3):034105. doi: 10.1063/1.2816557. J Chem Phys. 2008. PMID: 18205486
-
Overview of Methionine Adenosyltransferase 2A (MAT2A) as an Anticancer Target: Structure, Function, and Inhibitors.J Med Chem. 2022 Jul 28;65(14):9531-9547. doi: 10.1021/acs.jmedchem.2c00395. Epub 2022 Jul 7. J Med Chem. 2022. PMID: 35796517 Review.
-
Genetic modification and bioprocess optimization for S-Adenosyl-L-methionine biosynthesis.Subcell Biochem. 2012;64:327-41. doi: 10.1007/978-94-007-5055-5_16. Subcell Biochem. 2012. PMID: 23080258 Review.
Cited by
-
Transition Path Sampling Study of Engineered Enzymes That Catalyze the Morita-Baylis-Hillman Reaction: Why Is Enzyme Design so Difficult?J Chem Inf Model. 2024 Mar 25;64(6):2101-2111. doi: 10.1021/acs.jcim.4c00045. Epub 2024 Mar 7. J Chem Inf Model. 2024. PMID: 38451822 Free PMC article.
-
CHARMM-GUI EnzyDocker for Protein-Ligand Docking of Multiple Reactive States along a Reaction Coordinate in Enzymes.J Chem Theory Comput. 2025 Feb 25;21(4):2118-2128. doi: 10.1021/acs.jctc.4c01691. Epub 2025 Feb 14. J Chem Theory Comput. 2025. PMID: 39950957 Free PMC article.
-
Myosin-Catalyzed ATP Hydrolysis in the Presence of Disease-Causing Mutations: Mavacamten as a Way to Repair Mechanism.J Phys Chem B. 2024 May 16;128(19):4716-4727. doi: 10.1021/acs.jpcb.4c01601. Epub 2024 May 6. J Phys Chem B. 2024. PMID: 38708944 Free PMC article.
-
Perspective: Path Sampling Methods Applied to Enzymatic Catalysis.J Chem Theory Comput. 2022 Nov 8;18(11):6397-6406. doi: 10.1021/acs.jctc.2c00734. Epub 2022 Oct 28. J Chem Theory Comput. 2022. PMID: 36305863 Free PMC article. Review.
-
Directed Evolution's Selective Use of Quantum Tunneling in Designed Enzymes─A Combined Theoretical and Experimental Study.J Phys Chem B. 2025 Feb 6;129(5):1555-1562. doi: 10.1021/acs.jpcb.4c08169. Epub 2025 Jan 28. J Phys Chem B. 2025. PMID: 39874479
References
-
- Karplus M; McCammon AJ Molecular dynamics simulations of biomolecules. Nat. Struct. Mol. Biol 2002, 9, 646–652. - PubMed
-
- Schramm VL Enzymatic transition states and transition state analog design. Annu. Rev. Biochem 1998, 67, 693–720. - PubMed
-
- Zhang X; Houk KN Why Enzymes Are Proficient Catalysts: Beyond the Pauling Paradigm. Acc. Chem. Res 2005, 38, 379–385. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

