Gene expression and functional analysis of Aha1a and Aha1b in stress response in zebrafish
- PMID: 35830921
- PMCID: PMC10306239
- DOI: 10.1016/j.cbpb.2022.110777
Gene expression and functional analysis of Aha1a and Aha1b in stress response in zebrafish
Abstract
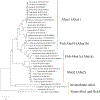
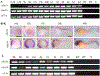
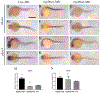
Activator of heat shock protein 90 (hsp90) ATPase (Aha1) is a Hsp90 co-chaperone required for Hsp90 ATPase activation. Aha1 is essential for yeast survival and muscle development in C. elegans under elevated temperature and hsp90-deficeiency induced stress conditions. The roles of Aha1 in vertebrates are poorly understood. Here, we characterized the expression and function of Aha1 in zebrafish. We showed that zebrafish genome contains two aha1 genes, aha1a and aha1b, that show distinct patterns of expression during development. Under the normal physiological conditions, aha1a is primarily expressed in skeletal muscle cells of zebrafish embryos, while aha1b is strongly expressed in the head region. aha1a and aha1b expression increased dramatically in response to heat shock induced stress. In addition, Aha1a-GFP fusion protein exhibited a dynamic translocation in muscle cells in response to heat shock. Moreover, upregulation of aha1 expression was also observed in hsp90a1 knockdown embryos that showed a muscle defect. Genetic studies demonstrated that knockout of aha1a, aha1b or both had no detectable effect on embryonic development, survival, and growth in zebrafish. The aha1a and aha1b mutant embryos showed normal muscle development and stress response in response to heat shock. Single or double aha1a and aha1b mutants could grow into normal reproductive adults with normal skeletal muscle structure and morphology compared with wild type control. Together, data from these studies indicate that Aha1a and Aha1b are involved in stress response. However, they are dispensable in zebrafish embryonic development, growth, and survival.
Keywords: Aha1(activator of Hsp90 ATPase); Heat shock protein; Heat shock response; Myosin chaperone.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures
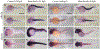



Similar articles
-
Hsf1 is essential for proteotoxic stress response in smyd1b-deficient embryos and fish survival under heat shock.FASEB J. 2025 Jan 15;39(1):e70283. doi: 10.1096/fj.202401875R. FASEB J. 2025. PMID: 39760245
-
A novel C-terminal homologue of Aha1 co-chaperone binds to heat shock protein 90 and stimulates its ATPase activity in Entamoeba histolytica.J Mol Biol. 2014 Apr 17;426(8):1786-98. doi: 10.1016/j.jmb.2014.01.008. Epub 2014 Jan 29. J Mol Biol. 2014. PMID: 24486610
-
p53 protein regulates Hsp90 ATPase activity and thereby Wnt signaling by modulating Aha1 expression.J Biol Chem. 2014 Mar 7;289(10):6513-6525. doi: 10.1074/jbc.M113.532523. Epub 2014 Jan 22. J Biol Chem. 2014. Retraction in: J Biol Chem. 2020 Jan 3;295(1):289. doi: 10.1074/jbc.W119.012134. PMID: 24451373 Free PMC article. Retracted.
-
Aha-type co-chaperones: the alpha or the omega of the Hsp90 ATPase cycle?Biol Chem. 2020 Mar 26;401(4):423-434. doi: 10.1515/hsz-2019-0341. Biol Chem. 2020. PMID: 31782942 Review.
-
p23 and Aha1: Distinct Functions Promote Client Maturation.Subcell Biochem. 2023;101:159-187. doi: 10.1007/978-3-031-14740-1_6. Subcell Biochem. 2023. PMID: 36520307 Review.
Cited by
-
Hsf1 is essential for proteotoxic stress response in smyd1b-deficient embryos and fish survival under heat shock.FASEB J. 2025 Jan 15;39(1):e70283. doi: 10.1096/fj.202401875R. FASEB J. 2025. PMID: 39760245
-
Genome-Wide Identification, Molecular Characterization, and Expression Analysis of the HSP70 and HSP90 Gene Families in Thamnaconus septentrionalis.Int J Mol Sci. 2024 May 24;25(11):5706. doi: 10.3390/ijms25115706. Int J Mol Sci. 2024. PMID: 38891896 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

