A SOX17-PDGFB signaling axis regulates aortic root development
- PMID: 35831318
- PMCID: PMC9279414
- DOI: 10.1038/s41467-022-31815-1
A SOX17-PDGFB signaling axis regulates aortic root development
Abstract
Developmental etiologies causing complex congenital aortic root abnormalities are unknown. Here we show that deletion of Sox17 in aortic root endothelium in mice causes underdeveloped aortic root leading to a bicuspid aortic valve due to the absence of non-coronary leaflet and mispositioned left coronary ostium. The respective defects are associated with reduced proliferation of non-coronary leaflet mesenchyme and aortic root smooth muscle derived from the second heart field cardiomyocytes. Mechanistically, SOX17 occupies a Pdgfb transcriptional enhancer to promote its transcription and Sox17 deletion inhibits the endothelial Pdgfb transcription and PDGFB growth signaling to the non-coronary leaflet mesenchyme. Restoration of PDGFB in aortic root endothelium rescues the non-coronary leaflet and left coronary ostium defects in Sox17 nulls. These data support a SOX17-PDGFB axis underlying aortic root development that is critical for aortic valve and coronary ostium patterning, thereby informing a potential shared disease mechanism for concurrent anomalous aortic valve and coronary arteries.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


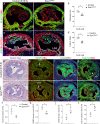

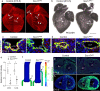
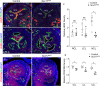

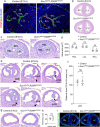

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

