CD8 lymphocytes mitigate HIV-1 persistence in lymph node follicular helper T cells during hyperacute-treated infection
- PMID: 35831418
- PMCID: PMC9279299
- DOI: 10.1038/s41467-022-31692-8
CD8 lymphocytes mitigate HIV-1 persistence in lymph node follicular helper T cells during hyperacute-treated infection
Abstract
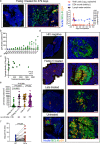
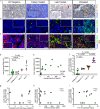
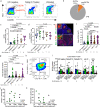
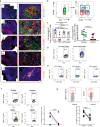
HIV persistence in tissue sites despite ART is a major barrier to HIV cure. Detailed studies of HIV-infected cells and immune responses in native lymph node tissue environment is critical for gaining insight into immune mechanisms impacting HIV persistence and clearance in tissue sanctuary sites. We compared HIV persistence and HIV-specific T cell responses in lymph node biopsies obtained from 14 individuals who initiated therapy in Fiebig stages I/II, 5 persons treated in Fiebig stages III-V and 17 late treated individuals who initiated ART in Fiebig VI and beyond. Using multicolor immunofluorescence staining and in situ hybridization, we detect HIV RNA and/or protein in 12 of 14 Fiebig I/II treated persons on suppressive therapy for 1 to 55 months, and in late treated persons with persistent antigens. CXCR3+ T follicular helper cells harbor the greatest amounts of gag mRNA transcripts. Notably, HIV-specific CD8+ T cells responses are associated with lower HIV antigen burden, suggesting that these responses may contribute to HIV suppression in lymph nodes during therapy. These results reveal HIV persistence despite the initiation of ART in hyperacute infection and highlight the contribution of virus-specific responses to HIV suppression in tissue sanctuaries during suppressive ART.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Human Immunodeficiency Virus Persistence and T-Cell Activation in Blood, Rectal, and Lymph Node Tissue in Human Immunodeficiency Virus-Infected Individuals Receiving Suppressive Antiretroviral Therapy.J Infect Dis. 2017 Mar 15;215(6):911-919. doi: 10.1093/infdis/jix039. J Infect Dis. 2017. PMID: 28453847 Free PMC article.
-
PD-1(+) and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals.Nat Med. 2016 Jul;22(7):754-61. doi: 10.1038/nm.4113. Epub 2016 May 30. Nat Med. 2016. PMID: 27239760
-
CD32+ and PD-1+ Lymph Node CD4 T Cells Support Persistent HIV-1 Transcription in Treated Aviremic Individuals.J Virol. 2018 Sep 26;92(20):e00901-18. doi: 10.1128/JVI.00901-18. Print 2018 Oct 15. J Virol. 2018. PMID: 29976671 Free PMC article.
-
The B-Cell Follicle in HIV Infection: Barrier to a Cure.Front Immunol. 2018 Jan 25;9:20. doi: 10.3389/fimmu.2018.00020. eCollection 2018. Front Immunol. 2018. PMID: 29422894 Free PMC article. Review.
-
Does B Cell Follicle Exclusion of CD8+ T Cells Make Lymph Nodes Sanctuaries of HIV Replication?Front Immunol. 2019 Oct 9;10:2362. doi: 10.3389/fimmu.2019.02362. eCollection 2019. Front Immunol. 2019. PMID: 31649673 Free PMC article. Review.
Cited by
-
Early treatment and PD1 inhibition enhance HIV-specific functionality of follicular CD8+ T cells.JCI Insight. 2025 Apr 8;10(7):e180309. doi: 10.1172/jci.insight.180309. JCI Insight. 2025. PMID: 40197363 Free PMC article.
-
Immune targeting of HIV-1 reservoir cells: a path to elimination strategies and cure.Nat Rev Microbiol. 2024 Jun;22(6):328-344. doi: 10.1038/s41579-024-01010-8. Epub 2024 Feb 9. Nat Rev Microbiol. 2024. PMID: 38337034 Free PMC article. Review.
-
Viral Persistence in the Gut-Associated Lymphoid Tissue and Barriers to HIV Cure.AIDS Res Hum Retroviruses. 2023 Dec 13;40(1):54-65. doi: 10.1089/AID.2022.0180. Online ahead of print. AIDS Res Hum Retroviruses. 2023. PMID: 37450338 Free PMC article.
-
Activation of CXCR3+ Tfh cells and B cells in lymph nodes during acute HIV-1 infection correlates with HIV-specific antibody development.J Virol. 2025 Mar 18;99(3):e0153224. doi: 10.1128/jvi.01532-24. Epub 2025 Feb 11. J Virol. 2025. PMID: 39932316 Free PMC article.
-
HIV-SEQ REVEALS GLOBAL HOST GENE EXPRESSION DIFFERENCES BETWEEN HIV-TRANSCRIBING CELLS FROM VIREMIC AND SUPPRESSED PEOPLE WITH HIV.bioRxiv [Preprint]. 2024 Dec 20:2024.12.17.629023. doi: 10.1101/2024.12.17.629023. bioRxiv. 2024. PMID: 39763963 Free PMC article. Preprint.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

