Assessing the response to genomic selection by simulation
- PMID: 35831462
- PMCID: PMC9325815
- DOI: 10.1007/s00122-022-04157-1
Assessing the response to genomic selection by simulation
Abstract
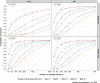
We propose a simulation approach to compute response to genomic selection on a multi-environment framework to provide breeders the number of entries that need to be selected from the population to have a defined probability of selecting the truly best entry from the population and the probability of obtaining the truly best entries when some top-ranked entries are selected. The goal of any plant breeding program is to maximize genetic gain for traits of interest. In classical quantitative genetics, the genetic gain can be obtained from what is known as "Breeder's equation". In the past, only phenotypic data were used to compute the genetic gain. The advent of genomic prediction (GP) has opened the door to the utilization of dense markers for estimating genomic breeding values or GBV. The salient feature of GP is the possibility to carry out genomic selection with the assistance of the kinship matrix, hence improving the prediction accuracy and accelerating the breeding cycle. However, estimates of GBV as such do not provide the full information on the number of entries to be selected as in the classical response to selection. In this paper, we use simulation, based on a fitted mixed model for GP in a multi-environmental framework, to answer two typical questions of a plant breeder: (1) How many entries need to be selected to have a defined probability of selecting the truly best entry from the population; (2) what is the probability of obtaining the truly best entries when some top-ranked entries are selected.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures



References
-
- Astle W, Balding DJ. Population structure and cryptic relatedness in genetic association studies. Stat Sci. 2009;24:451–471. doi: 10.1214/09-STS307. - DOI
-
- Buntaran H, Piepho H-P, Schmidt P, Rydén J, Halling M, Forkman J. Cross-validation of stagewise mixed-model analysis of Swedish variety trials with winter wheat and spring barley. Crop Sci. 2020;60:2221–2240. doi: 10.1002/csc2.20177. - DOI
-
- Butler DG, Cullis B, Gilmour A, Gogel BJ, Thompson R. ASReml-R reference manual, version 4. Wollongong: University of Wollongong; 2017.
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

