Redefining GBA gene structure unveils the ability of Cap-independent, IRES-dependent gene regulation
- PMID: 35831491
- PMCID: PMC9279297
- DOI: 10.1038/s42003-022-03577-5
Redefining GBA gene structure unveils the ability of Cap-independent, IRES-dependent gene regulation
Abstract
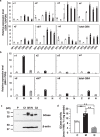
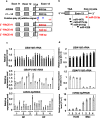
Glucosylceramide is the primary molecule of glycosphingolipids, and its metabolic regulation is crucial for life. Defects in the catabolizing enzyme, glucocerebrosidase (GCase), cause a lysosomal storage disorder known as Gaucher disease. However, the genetic regulation of GCase has not been fully understood. Here we show the redefined structure of the GCase coding gene (GBA), and clarify the regulatory mechanisms of its transcription and translation. First, alternative uses of the two GBA gene promoters were identified in fibroblasts and HL60-derived macrophages. Intriguingly, both GBA transcripts and GCase activities were induced in macrophages but not in neutrophils. Second, we observed cap-independent translation occurs via unique internal ribosome entry site activities in first promoter-driven GBA transcripts. Third, the reciprocal expression was observed in GBA and miR22-3p versus GBAP1 transcripts before and after HL60-induced macrophage differentiation. Nevertheless, these findings clearly demonstrate novel cell-type-specific GBA gene expression regulatory mechanisms, providing new insights into GCase biology.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
Potential Binding Sites of Pharmacological Chaperone NCGC00241607 on Mutant β-Glucocerebrosidase and Its Efficacy on Patient-Derived Cell Cultures in Gaucher and Parkinson's Disease.Int J Mol Sci. 2023 May 22;24(10):9105. doi: 10.3390/ijms24109105. Int J Mol Sci. 2023. PMID: 37240451 Free PMC article.
-
The GBAP1 pseudogene acts as a ceRNA for the glucocerebrosidase gene GBA by sponging miR-22-3p.Sci Rep. 2017 Oct 5;7(1):12702. doi: 10.1038/s41598-017-12973-5. Sci Rep. 2017. PMID: 28983119 Free PMC article.
-
Neurological effects of glucocerebrosidase gene mutations.Eur J Neurol. 2019 Mar;26(3):388-e29. doi: 10.1111/ene.13837. Epub 2018 Dec 13. Eur J Neurol. 2019. PMID: 30315684 Free PMC article. Review.
-
Ambroxol increases glucocerebrosidase (GCase) activity and restores GCase translocation in primary patient-derived macrophages in Gaucher disease and Parkinsonism.Parkinsonism Relat Disord. 2021 Mar;84:112-121. doi: 10.1016/j.parkreldis.2021.02.003. Epub 2021 Feb 10. Parkinsonism Relat Disord. 2021. PMID: 33609962
-
Cross-talks among GBA mutations, glucocerebrosidase, and α-synuclein in GBA-associated Parkinson's disease and their targeted therapeutic approaches: a comprehensive review.Transl Neurodegener. 2021 Jan 15;10(1):4. doi: 10.1186/s40035-020-00226-x. Transl Neurodegener. 2021. PMID: 33446243 Free PMC article. Review.
Cited by
-
Clinical, mechanistic, biomarker, and therapeutic advances in GBA1-associated Parkinson's disease.Transl Neurodegener. 2024 Sep 12;13(1):48. doi: 10.1186/s40035-024-00437-6. Transl Neurodegener. 2024. PMID: 39267121 Free PMC article. Review.
-
The Consequences of GBA Deficiency in the Autophagy-Lysosome System in Parkinson's Disease Associated with GBA.Cells. 2023 Jan 3;12(1):191. doi: 10.3390/cells12010191. Cells. 2023. PMID: 36611984 Free PMC article. Review.
-
GBA1 Gene Mutations in α-Synucleinopathies-Molecular Mechanisms Underlying Pathology and Their Clinical Significance.Int J Mol Sci. 2023 Jan 20;24(3):2044. doi: 10.3390/ijms24032044. Int J Mol Sci. 2023. PMID: 36768367 Free PMC article. Review.
-
Obstacles to Early Diagnosis of Gaucher Disease.Ther Clin Risk Manag. 2025 Jan 25;21:93-101. doi: 10.2147/TCRM.S388266. eCollection 2025. Ther Clin Risk Manag. 2025. PMID: 39882275 Free PMC article. Review.
-
Classification and Genotype-Phenotype Relationships of GBA1 Variants: MDSGene Systematic Review.Mov Disord. 2025 Apr;40(4):605-618. doi: 10.1002/mds.30141. Epub 2025 Feb 10. Mov Disord. 2025. PMID: 39927608 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

