Retrograde movements determine effective stem cell numbers in the intestine
- PMID: 35831497
- PMCID: PMC7614894
- DOI: 10.1038/s41586-022-04962-0
Retrograde movements determine effective stem cell numbers in the intestine
Abstract
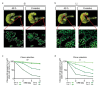
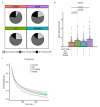
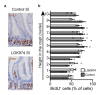
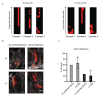
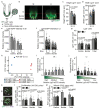
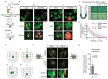
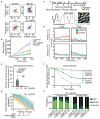
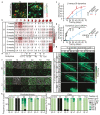
The morphology and functionality of the epithelial lining differ along the intestinal tract, but tissue renewal at all sites is driven by stem cells at the base of crypts1-3. Whether stem cell numbers and behaviour vary at different sites is unknown. Here we show using intravital microscopy that, despite similarities in the number and distribution of proliferative cells with an Lgr5 signature in mice, small intestinal crypts contain twice as many effective stem cells as large intestinal crypts. We find that, although passively displaced by a conveyor-belt-like upward movement, small intestinal cells positioned away from the crypt base can function as long-term effective stem cells owing to Wnt-dependent retrograde cellular movement. By contrast, the near absence of retrograde movement in the large intestine restricts cell repositioning, leading to a reduction in effective stem cell number. Moreover, after suppression of the retrograde movement in the small intestine, the number of effective stem cells is reduced, and the rate of monoclonal conversion of crypts is accelerated. Together, these results show that the number of effective stem cells is determined by active retrograde movement, revealing a new channel of stem cell regulation that can be experimentally and pharmacologically manipulated.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures






Comment in
-
Relocation sustains intestinal stem-cell numbers.Nature. 2022 Jul;607(7919):451-452. doi: 10.1038/d41586-022-01596-0. Nature. 2022. PMID: 35831566 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

