ZBTB46 defines and regulates ILC3s that protect the intestine
- PMID: 35831503
- PMCID: PMC9528687
- DOI: 10.1038/s41586-022-04934-4
ZBTB46 defines and regulates ILC3s that protect the intestine
Abstract
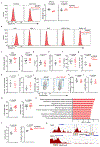
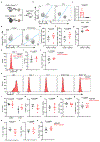
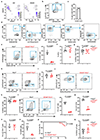
RORγt is a lineage-specifying transcription factor that is expressed by immune cells that are enriched in the gastrointestinal tract and promote immunity, inflammation and tissue homeostasis1-15. However, fundamental questions remain with regard to the cellular heterogeneity among these cell types, the mechanisms that control protective versus inflammatory properties and their functional redundancy. Here we define all RORγt+ immune cells in the intestine at single-cell resolution and identify a subset of group 3 innate lymphoid cells (ILC3s) that expresses ZBTB46, a transcription factor specifying conventional dendritic cells16-20. ZBTB46 is robustly expressed by CCR6+ lymphoid-tissue-inducer-like ILC3s that are developmentally and phenotypically distinct from conventional dendritic cells, and its expression is imprinted by RORγt, fine-tuned by microbiota-derived signals and increased by pro-inflammatory cytokines. ZBTB46 restrains the inflammatory properties of ILC3s, including the OX40L-dependent expansion of T helper 17 cells and the exacerbated intestinal inflammation that occurs after enteric infection. Finally, ZBTB46+ ILC3s are a major source of IL-22, and selective depletion of this population renders mice susceptible to enteric infection and associated intestinal inflammation. These results show that ZBTB46 is a transcription factor that is shared between conventional dendritic cells and ILC3s, and identify a cell-intrinsic function for ZBTB46 in restraining the pro-inflammatory properties of ILC3s and a non-redundant role for ZBTB46+ ILC3s in orchestrating intestinal health.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures











Comment in
-
ZBTB46+ ILC3s promote intestinal health.Nat Rev Gastroenterol Hepatol. 2022 Sep;19(9):555. doi: 10.1038/s41575-022-00672-0. Nat Rev Gastroenterol Hepatol. 2022. PMID: 35915158 No abstract available.
-
ZBTB46 in ILC3: shared transcriptional infrastructure defines gut-protective capabilities.Trends Immunol. 2022 Sep;43(9):690-692. doi: 10.1016/j.it.2022.07.008. Epub 2022 Aug 9. Trends Immunol. 2022. PMID: 35953346
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

