DNA damage and repair in age-related inflammation
- PMID: 35831609
- PMCID: PMC10106081
- DOI: 10.1038/s41577-022-00751-y
DNA damage and repair in age-related inflammation
Abstract
Genomic instability is an important driver of ageing. The accumulation of DNA damage is believed to contribute to ageing by inducing cell death, senescence and tissue dysfunction. However, emerging evidence shows that inflammation is another major consequence of DNA damage. Inflammation is a hallmark of ageing and the driver of multiple age-related diseases. Here, we review the evidence linking DNA damage, inflammation and ageing, highlighting how premature ageing syndromes are associated with inflammation. We discuss the mechanisms by which DNA damage induces inflammation, such as through activation of the cGAS-STING axis and NF-κB activation by ATM. The triggers for activation of these signalling cascades are the age-related accumulation of DNA damage, activation of transposons, cellular senescence and the accumulation of persistent R-loops. We also discuss how epigenetic changes triggered by DNA damage can lead to inflammation and ageing via redistribution of heterochromatin factors. Finally, we discuss potential interventions against age-related inflammation.
© 2022. Springer Nature Limited.
Conflict of interest statement
Conflict of Interest
VG is a consultant for Elysium, Centaura, Genflow Bio, and Do Not Age. Other authors declare no conflicting interests.
Figures


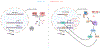


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

