Epigenetic modulators of B cell fate identified through coupled phenotype-transcriptome analysis
- PMID: 35831623
- PMCID: PMC9751284
- DOI: 10.1038/s41418-022-01037-5
Epigenetic modulators of B cell fate identified through coupled phenotype-transcriptome analysis
Abstract
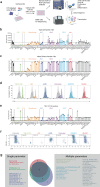
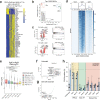
High-throughput methodologies are the cornerstone of screening approaches to identify novel compounds that regulate immune cell function. To identify novel targeted therapeutics to treat immune disorders and haematological malignancies, there is a need to integrate functional cellular information with the molecular mechanisms that regulate changes in immune cell phenotype. We facilitate this goal by combining quantitative methods for dissecting complex simultaneous cell phenotypic effects with genomic analysis. This combination strategy we term Multiplexed Analysis of Cells sequencing (MAC-seq), a modified version of Digital RNA with perturbation of Genes (DRUGseq). We applied MAC-seq to screen compounds that target the epigenetic machinery of B cells and assess altered humoral immunity by measuring changes in proliferation, survival, differentiation and transcription. This approach revealed that polycomb repressive complex 2 (PRC2) inhibitors promote antibody secreting cell (ASC) differentiation in both murine and human B cells in vitro. This is further validated using T cell-dependent immunization in mice. Functional dissection of downstream effectors of PRC2 using arrayed CRISPR screening uncovered novel regulators of B cell differentiation, including Mybl1, Myof, Gas7 and Atoh8. Together, our findings demonstrate that integrated phenotype-transcriptome analyses can be effectively combined with drug screening approaches to uncover the molecular circuitry that drives lymphocyte fate decisions.
© 2022. The Author(s).
Conflict of interest statement
The Johnstone laboratory (SJV and RWJ) has received project support from AstraZeneca, Roche, MecRx, and BMS. RWJ is a paid consultant and shareholder of MecRx.
Figures


References
-
- Trezise S, Nutt SL. The gene regulatory network controlling plasma cell function. Immunol Rev. 2021;303:23–34. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

