Comparative genomics of the plant-growth promoting bacterium Sphingobium sp. strain AEW4 isolated from the rhizosphere of the beachgrass Ammophila breviligulata
- PMID: 35831788
- PMCID: PMC9281055
- DOI: 10.1186/s12864-022-08738-8
Comparative genomics of the plant-growth promoting bacterium Sphingobium sp. strain AEW4 isolated from the rhizosphere of the beachgrass Ammophila breviligulata
Abstract
Background: The genus Sphingobium within the class Alpha-proteobacteria contains a small number of plant-growth promoting rhizobacteria (PGPR), although it is mostly comprised of organisms that play an important role in biodegradation and bioremediation in sediments and sandy soils. A Sphingobium sp. isolate was obtained from the rhizosphere of the beachgrass Ammophila breviligulata with a variety of plant growth-promoting properties and designated as Sphingobium sp. strain AEW4.
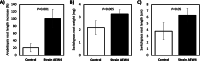
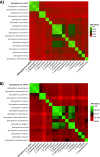
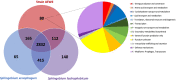
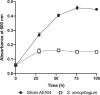
Results: Analysis of the 16S rRNA gene as well as full genome nucleotide and amino acid identities revealed that this isolate is most similar to Sphingobium xenophagum and Sphingobium hydrophobicum. Comparative genomics analyses indicate that the genome of strain AEW4 contains unique features that explain its relationship with a plant host as a PGPR, including pathways involved in monosaccharide utilization, fermentation pathways, iron sequestration, and resistance to osmotic stress. Many of these unique features are not broadly distributed across the genus. In addition, pathways involved in the metabolism of salicylate and catechol, phenyl acetate degradation, and DNA repair were also identified in this organism but not in most closely related organisms.
Conclusion: The genome of Sphingobium sp. strain AEW4 contains a number of distinctive features that are crucial to explain its role as a plant-growth promoting rhizobacterium, and comparative genomics analyses support its classification as a relevant Sphingobium strain involved in plant growth promotion of beachgrass and other plants.
Keywords: Beachgrass; Comparative genomics; PGPR; Rhizobacterium; Sphingobium.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Takeuchi M, Hamana K, Hiraishi A. Proposal of the genus Sphingomonas sensu stricto and three new genera, Sphingobium, Novosphingobium and Sphingopyxis, on the basis of phylogenetic and chemotaxonomic analyses. Int J Syst Evol Microbiol. 2001;51:1405–1417. doi: 10.1099/00207713-51-4-1405. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

