Integrated analysis of Wnt signalling system component gene expression
- PMID: 35831952
- PMCID: PMC9481969
- DOI: 10.1242/dev.200312
Integrated analysis of Wnt signalling system component gene expression
Abstract
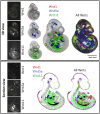
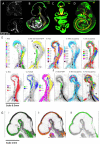
Wnt signalling controls patterning and differentiation across many tissues and organs of the developing embryo through temporally and spatially restricted expression of multi-gene families encoding ligands, receptors, pathway modulators and intracellular components. Here, we report an integrated analysis of key genes in the 3D space of the mouse embryo across multiple stages of development. We applied a method for 3D/3D image transformation to map all gene expression patterns to a single reference embryo for each stage, providing both visual analysis and volumetric mapping allowing computational methods to interrogate the combined expression patterns. We identify territories where multiple Wnt and Fzd genes are co-expressed and cross-compare all patterns, including all seven Wnt paralogous gene pairs. The comprehensive analysis revealed regions in the embryo where no Wnt or Fzd gene expression is detected, and where single Wnt genes are uniquely expressed. This work provides insight into a previously unappreciated level of organisation of expression patterns, as well as presenting a resource that can be utilised further by the research community for whole-system analysis.
Keywords: 3D imaging; Computational analysis; Integrated analysis; Wnt signalling.
© 2022. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

