Genome-wide local ancestry and evidence for mitonuclear coadaptation in African hybrid cattle populations
- PMID: 35832892
- PMCID: PMC9272374
- DOI: 10.1016/j.isci.2022.104672
Genome-wide local ancestry and evidence for mitonuclear coadaptation in African hybrid cattle populations
Abstract
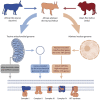
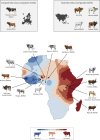
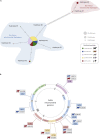
The phenotypic diversity of African cattle reflects adaptation to a wide range of agroecological conditions, human-mediated selection preferences, and complex patterns of admixture between the humpless Bos taurus (taurine) and humped Bos indicus (zebu) subspecies, which diverged 150-500 thousand years ago. Despite extensive admixture, all African cattle possess taurine mitochondrial haplotypes, even populations with significant zebu biparental and male uniparental nuclear ancestry. This has been interpreted as the result of human-mediated dispersal ultimately stemming from zebu bulls imported from South Asia during the last three millennia. Here, we assess whether ancestry at mitochondrially targeted nuclear genes in African admixed cattle is impacted by mitonuclear functional interactions. Using high-density SNP data, we find evidence for mitonuclear coevolution across hybrid African cattle populations with a significant increase of taurine ancestry at mitochondrially targeted nuclear genes. Our results, therefore, support the hypothesis of incompatibility between the taurine mitochondrial genome and the zebu nuclear genome.
Keywords: Biological sciences; Ecology; Evolutionary biology; Genetics; Genomics.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interest.
Figures


Similar articles
-
Selection Increases Mitonuclear DNA Discordance but Reconciles Incompatibility in African Cattle.Mol Biol Evol. 2025 Feb 3;42(2):msaf039. doi: 10.1093/molbev/msaf039. Mol Biol Evol. 2025. PMID: 39921600 Free PMC article.
-
The patterns of admixture, divergence, and ancestry of African cattle populations determined from genome-wide SNP data.BMC Genomics. 2020 Dec 7;21(1):869. doi: 10.1186/s12864-020-07270-x. BMC Genomics. 2020. PMID: 33287702 Free PMC article.
-
Mitonuclear incompatibility as a hidden driver behind the genome ancestry of African admixed cattle.BMC Biol. 2022 Jan 17;20(1):20. doi: 10.1186/s12915-021-01206-x. BMC Biol. 2022. PMID: 35039029 Free PMC article.
-
Genomic clues of the evolutionary history of Bos indicus cattle.Anim Genet. 2019 Dec;50(6):557-568. doi: 10.1111/age.12836. Epub 2019 Sep 2. Anim Genet. 2019. PMID: 31475748 Review.
-
Pre-pubertal and postpartum anestrus in tropical Zebu cattle.Anim Reprod Sci. 2004 Jul;82-83:373-87. doi: 10.1016/j.anireprosci.2004.05.006. Anim Reprod Sci. 2004. PMID: 15271467 Review.
Cited by
-
Selection Increases Mitonuclear DNA Discordance but Reconciles Incompatibility in African Cattle.Mol Biol Evol. 2025 Feb 3;42(2):msaf039. doi: 10.1093/molbev/msaf039. Mol Biol Evol. 2025. PMID: 39921600 Free PMC article.
-
Genetic Structure Analysis of 155 Transboundary and Local Populations of Cattle (Bos taurus, Bos indicus and Bos grunniens) Based on STR Markers.Int J Mol Sci. 2023 Mar 6;24(5):5061. doi: 10.3390/ijms24055061. Int J Mol Sci. 2023. PMID: 36902492 Free PMC article.
-
Association Analysis Provides Insights into Plant Mitonuclear Interactions.Mol Biol Evol. 2024 Feb 1;41(2):msae028. doi: 10.1093/molbev/msae028. Mol Biol Evol. 2024. PMID: 38324417 Free PMC article.
-
Genomic insights into the population history and adaptive traits of Latin American Criollo cattle.R Soc Open Sci. 2024 Mar 27;11(3):231388. doi: 10.1098/rsos.231388. eCollection 2024 Mar. R Soc Open Sci. 2024. PMID: 38571912 Free PMC article.
-
Mapping restricted introgression across the genomes of admixed indigenous African cattle breeds.Genet Sel Evol. 2023 Dec 14;55(1):91. doi: 10.1186/s12711-023-00861-8. Genet Sel Evol. 2023. PMID: 38097935 Free PMC article.
References
-
- Bahbahani H., Tijjani A., Mukasa C., Wragg D., Almathen F., Nash O., Akpa G.N., Mbole-Kariuki M., Malla S., Woolhouse M., et al. Signatures of selection for environmental adaptation and zebu × taurine hybrid fitness in East African Shorthorn Zebu. Front. Genet. 2017;8:68. doi: 10.3389/fgene.2017.00068. - DOI - PMC - PubMed
-
- Barbato M., Hailer F., Upadhyay M., Del Corvo M., Colli L., Negrini R., Kim E.S., Crooijmans R.P.M.A., Sonstegard T., Ajmone-Marsan P. Adaptive introgression from indicine cattle into white cattle breeds from Central Italy. Sci. Rep. 2020;10:1279. doi: 10.1038/s41598-020-57880-4. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

