Immune- and Stemness-Related Genes Revealed by Comprehensive Analysis and Validation for Cancer Immunity and Prognosis and Its Nomogram in Lung Adenocarcinoma
- PMID: 35833114
- PMCID: PMC9271778
- DOI: 10.3389/fimmu.2022.829057
Immune- and Stemness-Related Genes Revealed by Comprehensive Analysis and Validation for Cancer Immunity and Prognosis and Its Nomogram in Lung Adenocarcinoma
Abstract
Objective: Lung adenocarcinoma (LUAD) is a familiar lung cancer with a very poor prognosis. This study investigated the immune- and stemness-related genes to develop model related with cancer immunity and prognosis in LUAD.
Method: The Cancer Genome Atlas (TCGA) was utilized for obtaining original transcriptome data and clinical information. Differential expression, prognostic value, and correlation with clinic parameter of mRNA stemness index (mRNAsi) were conducted in LUAD. Significant mRNAsi-related module and hub genes were screened using weighted gene coexpression network analysis (WGCNA). Meanwhile, immune-related differential genes (IRGs) were screened in LUAD. Stem cell index and immune-related differential genes (SC-IRGs) were screened and further developed to construct prognosis-related model and nomogram. Comprehensive analysis of hub genes and subgroups, involving enrichment in the subgroup [gene set enrichment analysis (GSEA)], gene mutation, genetic correlation, gene expression, immune, tumor mutation burden (TMB), and drug sensitivity, used bioinformatics and reverse transcription polymerase chain reaction (RT-PCR) for verification.
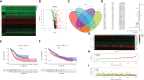
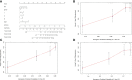
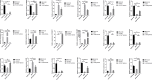
Results: Through difference analysis, mRNAsi of LUAD group was markedly higher than that of normal group. Clinical parameters (age, gender, and T staging) were ascertained to be highly relevant to mRNAsi. MEturquoise and MEblue were found to be the most significant modules (including positive and negative correlations) related to mRNAsi via WGCNA. The functions and pathways of the two mRNAsi-related modules were mainly enriched in tumorigenesis, development, and metastasis. Combining stem cell index-related differential genes and immune-related differential genes, 30 prognosis-related SC-IRGs were screened via Cox regression analysis. Then, 16 prognosis-related SC-IRGs were screened to construct a LASSO regression model at last. In addition, the model was successfully validated by using TCGA-LUAD and GSE68465, whereas c-index and the calibration curves were utilized to demonstrate the clinical value of our nomogram. Following the validation of the model, GSEA, immune cell correlation, TMB, clinical relevance, etc., have found significant difference in high- and low-risk groups, and 16-gene expression of the SC-IRG model also was tested by RT-PCR. ADRB2, ANGPTL4, BDNF, CBLC, CX3CR1, and IL3RA were found markedly different expression between the tumor and normal group.
Conclusion: The SC-IRG model and the prognostic nomogram could accurately predict LUAD survival. Our study used mRNAsi combined with immunity that may lay a foundation for the future research studies in LUAD.
Keywords: RT-PCR; cancer stem cell; immune; lung adenocarcinoma; muti-omics analysis; nomogram; stem cell index.
Copyright © 2022 Chen, Wang, Wang, Gui and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






Similar articles
-
Construction and validation of a prognostic model for stemness-related genes in lung adenocarcinoma.Transl Cancer Res. 2024 Mar 31;13(3):1351-1366. doi: 10.21037/tcr-23-1847. Epub 2024 Mar 11. Transl Cancer Res. 2024. PMID: 38617509 Free PMC article.
-
mRNAsi Index: Machine Learning in Mining Lung Adenocarcinoma Stem Cell Biomarkers.Genes (Basel). 2020 Feb 27;11(3):257. doi: 10.3390/genes11030257. Genes (Basel). 2020. PMID: 32121037 Free PMC article.
-
Cancer Stemness-Based Prognostic Immune-Related Gene Signatures in Lung Adenocarcinoma and Lung Squamous Cell Carcinoma.Front Endocrinol (Lausanne). 2021 Oct 21;12:755805. doi: 10.3389/fendo.2021.755805. eCollection 2021. Front Endocrinol (Lausanne). 2021. PMID: 34745015 Free PMC article.
-
Integrated genomic analysis of the stemness index signature of mRNA expression predicts lung adenocarcinoma prognosis and immune landscape.PeerJ. 2025 Feb 13;13:e18945. doi: 10.7717/peerj.18945. eCollection 2025. PeerJ. 2025. PMID: 39959839 Free PMC article.
-
Alternative Splicing in Lung Adenocarcinoma: From Bench to Bedside.Cancers (Basel). 2025 Apr 15;17(8):1329. doi: 10.3390/cancers17081329. Cancers (Basel). 2025. PMID: 40282505 Free PMC article. Review.
Cited by
-
Analysis and identification of mRNAsi‑related expression signatures via RNA sequencing in lung cancer.Oncol Lett. 2024 Sep 13;28(5):549. doi: 10.3892/ol.2024.14682. eCollection 2024 Nov. Oncol Lett. 2024. PMID: 39319211 Free PMC article.
-
Comprehensive Analysis Identifies Hyaluronan Mediated Motility Receptor and Cell Division Cycle 25C as Potential Prognostic Biomarkers in Head and Neck Squamous Cell Carcinoma.Cancer Control. 2024 Jan-Dec;31:10732748241287904. doi: 10.1177/10732748241287904. Cancer Control. 2024. PMID: 39323031 Free PMC article.
-
Molecular subgroup establishment and signature creation of lncRNAs associated with acetylation in lung adenocarcinoma.Aging (Albany NY). 2024 Jan 17;16(2):1276-1297. doi: 10.18632/aging.205407. Epub 2024 Jan 17. Aging (Albany NY). 2024. PMID: 38240708 Free PMC article.
-
Identification of cancer stemness and M2 macrophage-associated biomarkers in lung adenocarcinoma.Heliyon. 2023 Aug 16;9(9):e19114. doi: 10.1016/j.heliyon.2023.e19114. eCollection 2023 Sep. Heliyon. 2023. PMID: 37662825 Free PMC article.
-
Identification of specific prognostic markers for lung squamous cell carcinoma based on tumor progression, immune infiltration, and stem index.Front Immunol. 2023 Sep 29;14:1236444. doi: 10.3389/fimmu.2023.1236444. eCollection 2023. Front Immunol. 2023. PMID: 37841237 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

