FGF8-FGFR1 signaling regulates human GnRH neuron differentiation in a time- and dose-dependent manner
- PMID: 35833364
- PMCID: PMC9403748
- DOI: 10.1242/dmm.049436
FGF8-FGFR1 signaling regulates human GnRH neuron differentiation in a time- and dose-dependent manner
Abstract
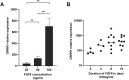
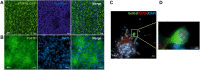
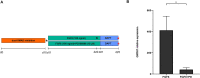
Fibroblast growth factor 8 (FGF8), acting through the fibroblast growth factor receptor 1 (FGFR1), has an important role in the development of gonadotropin-releasing hormone-expressing neurons (GnRH neurons). We hypothesized that FGF8 regulates differentiation of human GnRH neurons in a time- and dose-dependent manner via FGFR1. To investigate this further, human pluripotent stem cells were differentiated during 10 days of dual-SMAD inhibition into neural progenitor cells, followed either by treatment with FGF8 at different concentrations (25 ng/ml, 50 ng/ml or 100 ng/ml) for 10 days or by treatment with 100 ng/ml FGF8 for different durations (2, 4, 6 or 10 days); cells were then matured through DAPT-induced inhibition of Notch signaling for 5 days into GnRH neurons. FGF8 induced expression of GNRH1 in a dose-dependent fashion and the duration of FGF8 exposure correlated positively with gene expression of GNRH1 (P<0.05, Rs=0.49). However, cells treated with 100 ng/ml FGF8 for 2 days induced the expression of genes, such as FOXG1, ETV5 and SPRY2, and continued FGF8 treatment induced the dynamic expression of several other genes. Moreover, during exposure to FGF8, FGFR1 localized to the cell surface and its specific inhibition with the FGFR1 inhibitor PD166866 reduced expression of GNRH1 (P<0.05). In neurons, FGFR1 also localized to the nucleus. Our results suggest that dose- and time-dependent FGF8 signaling via FGFR1 is indispensable for human GnRH neuron ontogeny. This article has an associated First Person interview with the first author of the paper.
Keywords: FGF8; FGFR1; GnRH neuron; Transcriptome; hPSCs.
© 2022. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures




References
-
- Baron, O., Förthmann, B., Lee, Y.-W., Terranova, C., Ratzka, A., Stachowiak, E. K., Grothe, C., Claus, P. and Stachowiak, M. K. (2012). Cooperation of nuclear fibroblast growth factor receptor 1 and nurr1 offers new interactive mechanism in postmitotic development of mesencephalic dopaminergic neurons. J. Biol. Chem. 287, 19827-19840. 10.1074/jbc.M112.347831 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

