Adaptation by Type V-A and V-B CRISPR-Cas Systems Demonstrates Conserved Protospacer Selection Mechanisms Between Diverse CRISPR-Cas Types
- PMID: 35833800
- PMCID: PMC9419969
- DOI: 10.1089/crispr.2021.0150
Adaptation by Type V-A and V-B CRISPR-Cas Systems Demonstrates Conserved Protospacer Selection Mechanisms Between Diverse CRISPR-Cas Types
Abstract
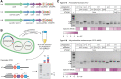
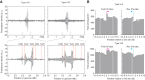
Adaptation of clustered regularly interspaced short palindromic repeats (CRISPR) arrays is a crucial process responsible for the unique, adaptive nature of CRISPR-Cas immune systems. The acquisition of new CRISPR spacers from mobile genetic elements has previously been studied for several types of CRISPR-Cas systems. In this study, we used a high-throughput sequencing approach to characterize CRISPR adaptation of the type V-A system from Francisella novicida and the type V-B system from Alicyclobacillus acidoterrestris. In contrast to other class 2 CRISPR-Cas systems, we found that for the type V-A and V-B systems, the Cas12 nucleases are dispensable for spacer acquisition, with only Cas1 and Cas2 (type V-A) or Cas4/1 and Cas2 (type V-B) being necessary and sufficient. Whereas the catalytic activity of Cas4 is not essential for adaptation, Cas4 activity is required for correct protospacer adjacent motif selection in both systems and for prespacer trimming in type V-A. In addition, we provide evidence for acquisition of RecBCD-produced DNA fragments by both systems, but with spacers derived from foreign DNA being incorporated preferentially over those derived from the host chromosome. Our work shows that several spacer acquisition mechanisms are conserved between diverse CRISPR-Cas systems, but also highlights unexpected nuances between similar systems that generally contribute to a bias of gaining immunity against invading genetic elements.
Conflict of interest statement
No competing financial interests exist.
Figures


Similar articles
-
Cas4 Nucleases Define the PAM, Length, and Orientation of DNA Fragments Integrated at CRISPR Loci.Mol Cell. 2018 Jun 7;70(5):814-824.e6. doi: 10.1016/j.molcel.2018.05.002. Epub 2018 Jun 7. Mol Cell. 2018. PMID: 29883605 Free PMC article.
-
Cas4 Nucleases Can Effect Specific Integration of CRISPR Spacers.J Bacteriol. 2019 May 22;201(12):e00747-18. doi: 10.1128/JB.00747-18. Print 2019 Jun 15. J Bacteriol. 2019. PMID: 30936372 Free PMC article.
-
Fidelity of prespacer capture and processing is governed by the PAM-mediated interactions of Cas1-2 adaptation complex in CRISPR-Cas type I-E system.J Biol Chem. 2019 Dec 27;294(52):20039-20053. doi: 10.1074/jbc.RA119.009438. Epub 2019 Nov 20. J Biol Chem. 2019. PMID: 31748409 Free PMC article.
-
CRISPR-Cas adaptation in Escherichia coli.Biosci Rep. 2023 Mar 31;43(3):BSR20221198. doi: 10.1042/BSR20221198. Biosci Rep. 2023. PMID: 36809461 Free PMC article. Review.
-
Creating memories: molecular mechanisms of CRISPR adaptation.Trends Biochem Sci. 2022 Jun;47(6):464-476. doi: 10.1016/j.tibs.2022.02.004. Epub 2022 Feb 28. Trends Biochem Sci. 2022. PMID: 35236570 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
