A joint PCR-based gene-targeting method using electroporation in the pathogenic fungus Trichosporon asahii
- PMID: 35834071
- PMCID: PMC9283638
- DOI: 10.1186/s13568-022-01431-9
A joint PCR-based gene-targeting method using electroporation in the pathogenic fungus Trichosporon asahii
Abstract
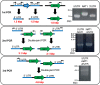
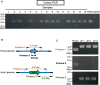
Trichosporon asahii is a pathogenic fungus that causes deep-seated fungal infections in immunocompromised patients. Established methods for generating gene-deficient T. asahii mutants exist, but the frequency of obtaining transformants by electroporation remains low. In the present study, we optimized the conditions for gene transfer by electroporation using a ku70 gene-deficient mutant with high recombination efficiency. Introducing a DNA fragment by electroporation into T. asahii cells on Sabouraud dextrose agar to generate a cnb1 gene-deficient mutant and incubating for 1 day led to the growth of approximately 100 transformants. When the incubation period was extended to 2 days or 5 days, however, only 2 or no transformants, respectively, were grown. The highest number of transformants was grown by electroporation when a square wave at 1.8 kV (9 kV/cm) was applied for 5 ms. In addition, the number of transformants increased with an increase in the length of the homologous region, and transformants did not grow when the homologous region was less than 500 base pairs. A DNA fragment was produced for deletion of the cnb1 gene by joint PCR, and the cnb1 gene-deficient mutant was obtained by introducing the DNA fragment by electroporation. These results indicate that DNA fragments produced by joint PCR can be used to generate gene-deficient mutants of T. asahii through gene transfer by electroporation.
Keywords: Electroporation; Gene targeting; Joint PCR; Mutant; Trichosporon asahii.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources

