Frequent detection but lack of infectivity of SARS-CoV-2 RNA in presymptomatic, infected blood donor plasma
- PMID: 35834347
- PMCID: PMC9435642
- DOI: 10.1172/JCI159876
Frequent detection but lack of infectivity of SARS-CoV-2 RNA in presymptomatic, infected blood donor plasma
Abstract
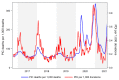
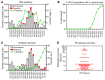
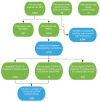
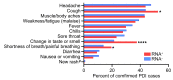
Respiratory viruses such as influenza do not typically cause viremia; however, SARS-CoV-2 has been detected in the blood of COVID-19 patients with mild and severe symptoms. Detection of SARS-CoV-2 in blood raises questions about its role in pathogenesis as well as transfusion safety concerns. Blood donor reports of symptoms or a diagnosis of COVID-19 after donation (post-donation information, PDI) preceded or coincided with increased general population COVID-19 mortality. Plasma samples from 2,250 blood donors who reported possible COVID-19-related PDI were tested for the presence of SARS-CoV-2 RNA. Detection of RNAemia peaked at 9%-15% of PDI donors in late 2020 to early 2021 and fell to approximately 4% after implementation of widespread vaccination in the population. RNAemic donors were 1.2- to 1.4-fold more likely to report cough or shortness of breath and 1.8-fold more likely to report change in taste or smell compared with infected donors without detectable RNAemia. No infectious virus was detected in plasma from RNAemic donors; inoculation of permissive cell lines produced less than 0.7-7 plaque-forming units (PFU)/mL and in susceptible mice less than 100 PFU/mL in RNA-positive plasma based on limits of detection in these models. These findings suggest that blood transfusions are highly unlikely to transmit SARS-CoV-2 infection.
Keywords: COVID-19; Clinical practice; Molecular diagnosis.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

