Profiling 26,000 Aplysia californica neurons by single cell mass spectrometry reveals neuronal populations with distinct neuropeptide profiles
- PMID: 35835221
- PMCID: PMC9396074
- DOI: 10.1016/j.jbc.2022.102254
Profiling 26,000 Aplysia californica neurons by single cell mass spectrometry reveals neuronal populations with distinct neuropeptide profiles
Abstract
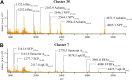
Neuropeptides are a chemically diverse class of cell-to-cell signaling molecules that are widely expressed throughout the central nervous system, often in a cell-specific manner. While cell-to-cell differences in neuropeptides is expected, it is often unclear how exactly neuropeptide expression varies among neurons. Here we created a microscopy-guided, high-throughput single cell matrix-assisted laser desorption/ionization mass spectrometry approach to investigate the neuropeptide heterogeneity of individual neurons in the central nervous system of the neurobiological model Aplysia californica, the California sea hare. In all, we analyzed more than 26,000 neurons from 18 animals and assigned 866 peptides from 66 prohormones by mass matching against an in silico peptide library generated from known Aplysia prohormones retrieved from the UniProt database. Louvain-Jaccard (LJ) clustering of mass spectra from individual neurons revealed 40 unique neuronal populations, or LJ clusters, each with a distinct neuropeptide profile. Prohormones and their related peptides were generally found in single cells from ganglia consistent with the prohormones' previously known ganglion localizations. Several LJ clusters also revealed the cellular colocalization of behaviorally related prohormones, such as an LJ cluster exhibiting achatin and neuropeptide Y, which are involved in feeding, and another cluster characterized by urotensin II, small cardiac peptide, sensorin A, and FRFa, which have shown activity in the feeding network or are present in the feeding musculature. This mass spectrometry-based approach enables the robust categorization of large cell populations based on single cell neuropeptide content and is readily adaptable to the study of a range of animals and tissue types.
Keywords: Aplysia californica; Louvain–Jaccard; central nervous system; mass spectrometry; neuron; neuropeptide; peptides; single cell.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures





References
-
- Strand F.L. In: Handbook of Comtemporary Neuropharmacology. Sibley D., Hanin I., Kuhar M., Skolnick P., editors. John Wiley & Sons; New York, NY: 2007. Neuropeptides; pp. 669–704.
-
- Hillebrand J.J.G., De Wied D., Adan R.A.H. Neuropeptides, food intake and body weight regulation: a hypothalamic focus. Peptides. 2002;23:2283–2306. - PubMed
-
- Xu P., Hall A.K. The role of activin in neuropeptide induction and pain sensation. Dev. Biol. 2006;299:303–309. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

