Pneumolysin suppresses the initial macrophage pro-inflammatory response to Streptococcus pneumoniae
- PMID: 35835695
- PMCID: PMC10497322
- DOI: 10.1111/imm.13546
Pneumolysin suppresses the initial macrophage pro-inflammatory response to Streptococcus pneumoniae
Abstract
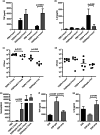
Published data for the Streptococcus pneumoniae virulence factor Pneumolysin (Ply) show contradictory effects on the host inflammatory response to infection. Ply has been shown to activate the inflammasome, but also can bind to MRC-1 resulting in suppression of dendritic cell inflammatory responses. We have used an in vitro infection model of human monocyte-derived macrophages (MDM), and a mouse model of pneumonia to clarify whether pro- or anti-inflammatory effects dominate the effects of Ply on the initial macrophage inflammatory response to S. pneumoniae, and the consequences during early lung infection. We found that infection with S. pneumoniae expressing Ply suppressed tumour necrosis factor (TNF) and interleukin-6 production by MDMs compared to cells infected with ply-deficient S. pneumoniae. This effect was independent of bacterial effects on cell death. Transcriptional analysis demonstrated S. pneumoniae expressing Ply caused a qualitatively similar but quantitatively lower MDM transcriptional response to S. pneumoniae compared to ply-deficient S. pneumoniae, with reduced expression of TNF and type I IFN inducible genes. Reduction of the MDM inflammatory response was prevented by inhibition of SOCS1. In the early lung infection mouse model, the TNF response to ply-deficient S. pneumoniae was enhanced and bacterial clearance increased compared to infection with wild-type S. pneumoniae. Overall, these data show Ply inhibits the initial macrophage inflammatory response to S. pneumoniae, probably mediated through SOCS1, and this was associated with improved immune evasion during early lung infection.
Keywords: Streptococcus pneumoniae; epithelial immunity; inflammation; pneumolysin.
© 2022 The Authors. Immunology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- O'Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria‐Knoll M, McCall N, et al. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet. 2009;374:893–902. - PubMed
-
- Dockrell DH, Marriott HM, Prince LR, Ridger VC, Ince PG, Hellewell PG, et al. Alveolar macrophage apoptosis contributes to pneumococcal clearance in a resolving model of pulmonary infection. J Immunol. 2003;171:5380–8. - PubMed
-
- Marriott HM, Gascoyne KA, Gowda R, Geary I, Nicklin MJH, Iannelli F, et al. Interleukin‐1β regulates CXCL8 release and influences disease outcome in response to Streptococcus pneumoniae, defining intercellular cooperation between pulmonary epithelial cells and macrophages. Infect Immun. 2012;80:1140–9. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

