In silico identification of two peptides with antibacterial activity against multidrug-resistant Staphylococcus aureus
- PMID: 35835775
- PMCID: PMC9283466
- DOI: 10.1038/s41522-022-00320-0
In silico identification of two peptides with antibacterial activity against multidrug-resistant Staphylococcus aureus
Abstract
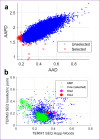
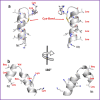
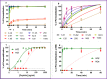
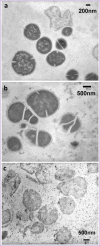
Here we report two antimicrobial peptides (AMPs), HG2 and HG4 identified from a rumen microbiome metagenomic dataset, with activity against multidrug-resistant (MDR) bacteria, especially methicillin-resistant Staphylococcus aureus (MRSA) strains, a major hospital and community-acquired pathogen. We employed the classifier model design to analyse, visualise, and interpret AMP activities. This approach allowed in silico discrimination of promising lead AMP candidates for experimental evaluation. The lead AMPs, HG2 and HG4, are fast-acting and show anti-biofilm and anti-inflammatory activities in vitro and demonstrated little toxicity to human primary cell lines. The peptides were effective in vivo within a Galleria mellonella model of MRSA USA300 infection. In terms of mechanism of action, HG2 and HG4 appear to interact with the cytoplasmic membrane of target cells and may inhibit other cellular processes, whilst preferentially binding to bacterial lipids over human cell lipids. Therefore, these AMPs may offer additional therapeutic templates for MDR bacterial infections.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Fnal Report and Recommendations (Review on Antimicrobial Resistance, 2016).
-
- Lewis, P. O., Heil, E. L., Covert, K. L. & Cluck, D. B. Treatment strategies for persistent methicillin-resistant Staphylococcus aureus bacteraemia. J. Clin. Pharm. Ther. 43, 614–625 10.1111/jcpt.12743 (2018). - PubMed
-
- WHO. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. (WHO, 2017).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

