Genetic regulation of OAS1 nonsense-mediated decay underlies association with COVID-19 hospitalization in patients of European and African ancestries
- PMID: 35835913
- PMCID: PMC9355882
- DOI: 10.1038/s41588-022-01113-z
Genetic regulation of OAS1 nonsense-mediated decay underlies association with COVID-19 hospitalization in patients of European and African ancestries
Abstract
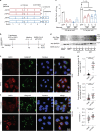
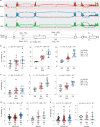
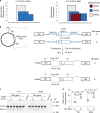
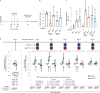
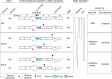
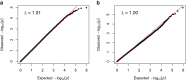
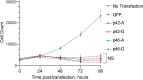
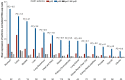
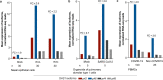
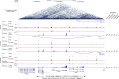
The chr12q24.13 locus encoding OAS1-OAS3 antiviral proteins has been associated with coronavirus disease 2019 (COVID-19) susceptibility. Here, we report genetic, functional and clinical insights into this locus in relation to COVID-19 severity. In our analysis of patients of European (n = 2,249) and African (n = 835) ancestries with hospitalized versus nonhospitalized COVID-19, the risk of hospitalized disease was associated with a common OAS1 haplotype, which was also associated with reduced severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) clearance in a clinical trial with pegIFN-λ1. Bioinformatic analyses and in vitro studies reveal the functional contribution of two associated OAS1 exonic variants comprising the risk haplotype. Derived human-specific alleles rs10774671-A and rs1131454 -A decrease OAS1 protein abundance through allele-specific regulation of splicing and nonsense-mediated decay (NMD). We conclude that decreased OAS1 expression due to a common haplotype contributes to COVID-19 severity. Our results provide insight into molecular mechanisms through which early treatment with interferons could accelerate SARS-CoV-2 clearance and mitigate against severe COVID-19.
© 2022. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
J.J.F. reports receiving research support unrelated to this work from Eiger BioPharmaceuticals. All other authors declare no competing interests.
Figures








Update of
-
Genetic regulation of OAS1 nonsense-mediated decay underlies association with risk of severe COVID-19.medRxiv [Preprint]. 2021 Jul 13:2021.07.09.21260221. doi: 10.1101/2021.07.09.21260221. medRxiv. 2021. Update in: Nat Genet. 2022 Aug;54(8):1103-1116. doi: 10.1038/s41588-022-01113-z. PMID: 34282422 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

