Prediction of the Mechanism of Sodium Butyrate against Radiation-Induced Lung Injury in Non-Small Cell Lung Cancer Based on Network Pharmacology and Molecular Dynamic Simulations and Molecular Dynamic Simulations
- PMID: 35837112
- PMCID: PMC9275827
- DOI: 10.3389/fonc.2022.809772
Prediction of the Mechanism of Sodium Butyrate against Radiation-Induced Lung Injury in Non-Small Cell Lung Cancer Based on Network Pharmacology and Molecular Dynamic Simulations and Molecular Dynamic Simulations
Abstract
Background: Radiation-induced lung injury (RILI) is a severe side effect of radiotherapy for non-small cell lung cancer (NSCLC) ,and one of the major hindrances to improve the efficacy of radiotherapy. Previous studies have confirmed that sodium butyrate (NaB) has potential of anti-radiation toxicity. However, the mechanism of the protective effect of NaB against RILI has not yet been clarified. This study aimed to explore the underlying protective mechanisms of NaB against RILI in NSCLC through network pharmacology, molecular docking, molecular dynamic simulations and in vivo experiments.
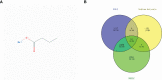
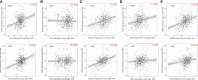
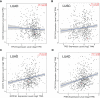
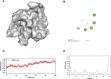
Methods: The predictive target genes of NaB were obtained from the PharmMapper database and the literature review. The involved genes of RILI and NSCLC were predicted using OMIM and GeneCards database. The intersectional genes of drug and disease were identified using the Venny tool and uploaded to the Cytoscape software to identify 5 core target genes of NaB associated with RILI. The correlations between the 5 core target genes and EGFR, PD-L1, immune infiltrates, chemokines and chemokine receptors were analyzed using TIMER 2.0, TIMER and TISIDB databases. We constructed the mechanism maps of the 3 key signaling pathways using the KEGG database based on the results of GO and KEGG analyses from Metascape database. The 5 core target genes and drug were docked using the AutoDock Vina tool and visualized using PyMOL software. GROMACS software was used to perform 100 ns molecular dynamics simulation. Irradiation-induced lung injury model in mice were established to assess the therapeutic effects of NaB.
Results: A total of 51 intersectional genes involved in NaB against RILI in NSCLC were identified. The 5 core target genes were AKT1, TP53, NOTCH1, SIRT1, and PTEN. The expressions of the 5 core target genes were significantly associated with EGFR, PD-L1, immune infiltrates, chemokines and chemokine receptors, respectively. The results from GO analysis of the 51 intersectional genes revealed that the biological processes were focused on the regulation of smooth muscle cell proliferation, oxidative stress and cell death, while the three key KEGG pathways were enriched in PI3K-Akt signal pathway, p53 signal pathway, and FOXO signal pathway. The docking of NaB with the 5 core target genes showed affinity and stability, especially AKT1. In vivo experiments showed that NaB treatment significantly protected mice from RILI, with reduced lung histological damage. In addition, NaB treatment significantly inhibited the PI3K/Akt signaling pathway.
Conclusions: NaB may protect patients from RILI in NSCLC through multiple target genes including AKT1, TP53, NOTCH1, SIRT1 and PTEN, with multiple signaling pathways involving, including PI3K-Akt pathway, p53 pathway, and FOXO pathways. Our findings effectively provide a feasible theoretical basis to further elucidate the mechanism of NaB in the treatment of RILI.
Keywords: molecular docking; molecular dynamics simulation; network pharmacology; non-small cell lung cancer; radiation-induced lung injury; signaling pathway; sodium butyrate; target gene.
Copyright © 2022 Zhang, Chen, Fan, Su, Liang and Jiang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures











Similar articles
-
Using Network Pharmacology and Molecular Docking to Explore the Mechanism of Shan Ci Gu (Cremastra appendiculata) Against Non-Small Cell Lung Cancer.Front Chem. 2021 Jun 9;9:682862. doi: 10.3389/fchem.2021.682862. eCollection 2021. Front Chem. 2021. PMID: 34178945 Free PMC article.
-
Network module analysis and molecular docking-based study on the mechanism of astragali radix against non-small cell lung cancer.BMC Complement Med Ther. 2023 Sep 28;23(1):345. doi: 10.1186/s12906-023-04148-9. BMC Complement Med Ther. 2023. PMID: 37770919 Free PMC article.
-
Mechanistic prediction and validation of Brevilin A Therapeutic effects in Lung Cancer.BMC Complement Med Ther. 2024 Jun 5;24(1):214. doi: 10.1186/s12906-024-04516-z. BMC Complement Med Ther. 2024. PMID: 38840248 Free PMC article.
-
Effects of Curcumin on Radiation/Chemotherapy-Induced Oral Mucositis: Combined Meta-Analysis, Network Pharmacology, Molecular Docking, and Molecular Dynamics Simulation.Curr Issues Mol Biol. 2024 Sep 20;46(9):10545-10569. doi: 10.3390/cimb46090625. Curr Issues Mol Biol. 2024. PMID: 39329977 Free PMC article. Review.
-
Cytological changes in radiation-induced lung injury.Life Sci. 2024 Dec 1;358:123188. doi: 10.1016/j.lfs.2024.123188. Epub 2024 Oct 29. Life Sci. 2024. PMID: 39481833 Review.
Cited by
-
Sodium butyrate supresses malignant human mast cell proliferation, downregulates expression of KIT and promotes differentiation.Front Allergy. 2023 Mar 10;4:1109717. doi: 10.3389/falgy.2023.1109717. eCollection 2023. Front Allergy. 2023. PMID: 36970068 Free PMC article.
-
Butyrate Metabolism-Related Gene Signature in Tumor Immune Microenvironment in Lung Adenocarcinoma: A Comprehensive Bioinformatics Study.Immun Inflamm Dis. 2024 Dec;12(12):e70087. doi: 10.1002/iid3.70087. Immun Inflamm Dis. 2024. PMID: 39641239 Free PMC article.
-
Early, non-invasive detection of radiation-induced lung injury using PET/CT by targeting CXCR4.Eur J Nucl Med Mol Imaging. 2024 Mar;51(4):1109-1120. doi: 10.1007/s00259-023-06517-5. Epub 2023 Nov 30. Eur J Nucl Med Mol Imaging. 2024. PMID: 38030744
-
Modification-specific Proteomic Analysis Reveals Cysteine S-Palmitoylation Involved in Esophageal Cancer Cell Radiation.ACS Omega. 2024 Dec 31;10(1):1541-1550. doi: 10.1021/acsomega.4c09353. eCollection 2025 Jan 14. ACS Omega. 2024. PMID: 39829482 Free PMC article.
-
Anticancer Effects of Gut Microbiota-Derived Short-Chain Fatty Acids in Cancers.J Microbiol Biotechnol. 2023 Jul 28;33(7):849-856. doi: 10.4014/jmb.2301.01031. Epub 2023 Mar 30. J Microbiol Biotechnol. 2023. PMID: 37100764 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

