Spatiotemporal Patterns of Substance P-Bound MRGPRX2 Reveal a Novel Connection Between Macropinosome Resolution and Secretory Granule Regeneration in Mast Cells
- PMID: 35837385
- PMCID: PMC9273857
- DOI: 10.3389/fimmu.2022.892239
Spatiotemporal Patterns of Substance P-Bound MRGPRX2 Reveal a Novel Connection Between Macropinosome Resolution and Secretory Granule Regeneration in Mast Cells
Abstract
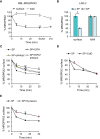
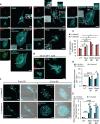
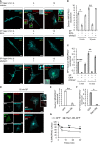
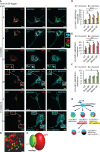
MRGPRX2, the human member of the MAS-related G protein coupled receptors (Mrgprs), serves as the cellular target of human mast cells (MCs) for innate ligands, including neuropeptides and antimicrobial peptides. In addition, MRGPRX2 also functions as the receptor for multiple FDA-approved drugs. As such, MRGPRX2 is a mediator of MC responses in neurogenic inflammation, host defense and pseudoallergy. We analyzed the spatiotemporal patterns of MRGPRX2 following its binding of the neuropeptide substance P (SP). Herein, we show that MRGPRX2 internalizes via both endocytosis and macropinocytosis, followed by its distribution between a perinuclear region and the secretory granules (SGs). Further, we show that MRGPRX2-containing macropinosomes undergo resolution by a mechanism that involves dynamin and LC3, giving rise to the incorporation of both LC3 and MRGPRX2 into the SGs. SP then promotes the acidification of the LC3-associated SGs, presumably by stimulating their fusion with lysosomes. Taken together, our results reveal a unique mode of MRGPRX2 trafficking that complements endocytosis and involves macropinocytosis, autophagic machinery-assisted macropinosome resolution and receptor delivery to the SGs.
Keywords: MRGPRX2; autophagy; endocytosis; macropinocytosis; mast cell; secretory granules; substance P.
Copyright © 2022 Lazki-Hagenbach, Kleeblatt, Ali and Sagi-Eisenberg.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
The Underlying Rab Network of MRGPRX2-Stimulated Secretion Unveils the Impact of Receptor Trafficking on Secretory Granule Biogenesis and Secretion.Cells. 2024 Jan 1;13(1):93. doi: 10.3390/cells13010093. Cells. 2024. PMID: 38201297 Free PMC article.
-
Emerging Roles for MAS-Related G Protein-Coupled Receptor-X2 in Host Defense Peptide, Opioid, and Neuropeptide-Mediated Inflammatory Reactions.Adv Immunol. 2017;136:123-162. doi: 10.1016/bs.ai.2017.06.002. Epub 2017 Jul 24. Adv Immunol. 2017. PMID: 28950944 Review.
-
Naturally Occurring Missense MRGPRX2 Variants Display Loss of Function Phenotype for Mast Cell Degranulation in Response to Substance P, Hemokinin-1, Human β-Defensin-3, and Icatibant.J Immunol. 2018 Jul 15;201(2):343-349. doi: 10.4049/jimmunol.1701793. Epub 2018 May 23. J Immunol. 2018. PMID: 29794017 Free PMC article.
-
Authentic and Ectopically Expressed MRGPRX2 Elicit Similar Mechanisms to Stimulate Degranulation of Mast Cells.Cells. 2021 Feb 12;10(2):376. doi: 10.3390/cells10020376. Cells. 2021. PMID: 33673037 Free PMC article.
-
Roles of Mas-related G protein-coupled receptor X2 on mast cell-mediated host defense, pseudoallergic drug reactions, and chronic inflammatory diseases.J Allergy Clin Immunol. 2016 Sep;138(3):700-710. doi: 10.1016/j.jaci.2016.04.051. Epub 2016 Jul 20. J Allergy Clin Immunol. 2016. PMID: 27448446 Free PMC article. Review.
Cited by
-
Inflammation and Organ Injury the Role of Substance P and Its Receptors.Int J Mol Sci. 2023 Mar 24;24(7):6140. doi: 10.3390/ijms24076140. Int J Mol Sci. 2023. PMID: 37047113 Free PMC article. Review.
-
p16Ink4a-induced senescence in cultured mast cells as a model for ageing reveals significant morphological and functional changes.Immun Ageing. 2024 Nov 11;21(1):77. doi: 10.1186/s12979-024-00478-5. Immun Ageing. 2024. PMID: 39529115 Free PMC article.
-
The Underlying Rab Network of MRGPRX2-Stimulated Secretion Unveils the Impact of Receptor Trafficking on Secretory Granule Biogenesis and Secretion.Cells. 2024 Jan 1;13(1):93. doi: 10.3390/cells13010093. Cells. 2024. PMID: 38201297 Free PMC article.
-
Biogenesis and homeostasis of mast cell lysosome related secretory granules.Front Cell Dev Biol. 2025 May 23;13:1603999. doi: 10.3389/fcell.2025.1603999. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40486912 Free PMC article. Review.
-
Substance P analogs devoid of key residues fail to activate human mast cells via MRGPRX2.Front Immunol. 2023 May 9;14:1155740. doi: 10.3389/fimmu.2023.1155740. eCollection 2023. Front Immunol. 2023. PMID: 37228611 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

