PtrWOX13A Promotes Wood Formation and Bioactive Gibberellins Biosynthesis in Populus trichocarpa
- PMID: 35837467
- PMCID: PMC9274204
- DOI: 10.3389/fpls.2022.835035
PtrWOX13A Promotes Wood Formation and Bioactive Gibberellins Biosynthesis in Populus trichocarpa
Abstract
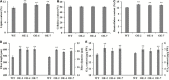
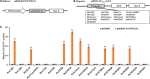
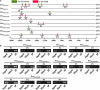
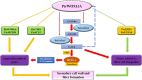
WUSCHEL-related homeobox (WOX) genes are plant-specific transcription factors (TFs) involved in multiple processes of plant development. However, there have hitherto no studies on the WOX TFs involved in secondary cell wall (SCW) formation been reported. In this study, we identified a Populus trichocarpa WOX gene, PtrWOX13A, which was predominantly expressed in SCW, and then characterized its functions through generating PtrWOX13A overexpression poplar transgenic lines; these lines exhibited not only significantly enhanced growth potential, but also remarkably increased SCW thicknesses, fiber lengths, and lignin and hemicellulose contents. However, no obvious change in cellulose content was observed. We revealed that PtrWOX13A directly activated its target genes through binding to two cis-elements, ATTGATTG and TTAATSS, in their promoter regions. The fact that PtrWOX13A responded to the exogenous GAs implies that it is responsive to GA homeostasis caused by GA inactivation and activation genes (e.g., PtrGA20ox4, PtrGA2ox1, and PtrGA3ox1), which were regulated by PtrWOX13A directly or indirectly. Since the master switch gene of SCW formation, PtrWND6A, and lignin biosynthesis regulator, MYB28, significantly increased in PtrWOX13A transgenic lines, we proposed that PtrWOX13A, as a higher hierarchy TF, participated in SCW formation through controlling the genes that are components of the known hierarchical transcription regulation network of poplar SCW formation, and simultaneously triggering a gibberellin-mediated signaling cascade. The discovery of PtrWOX13A predominantly expressed in SCW and its regulatory functions in the poplar wood formation has important implications for improving the wood quality of trees via genetic engineering.
Keywords: Populus trichocarpa; PtrWOX13A; bioactive gibberellins biosynthesis; coordinated regulation; wood formation.
Copyright © 2022 Zhang, Liu, Wang, Wang, Chen, Wang, Wei and Wei.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Characterization of a High Hierarchical Regulator, PtrGATA12, Functioning in Differentially Regulating Secondary Wall Component Biosynthesis in Populus trichocarpa.Front Plant Sci. 2021 Apr 21;12:657787. doi: 10.3389/fpls.2021.657787. eCollection 2021. Front Plant Sci. 2021. PMID: 33968111 Free PMC article.
-
PtrHAT22, as a higher hierarchy regulator, coordinately regulates secondary cell wall component biosynthesis in Populus trichocarpa.Plant Sci. 2022 Mar;316:111170. doi: 10.1016/j.plantsci.2021.111170. Epub 2021 Dec 30. Plant Sci. 2022. PMID: 35151454
-
WUSCHEL-RELATED HOMEOBOX genes are crucial for normal vascular organization and wood formation in poplar.Plant Sci. 2024 Sep;346:112138. doi: 10.1016/j.plantsci.2024.112138. Epub 2024 May 31. Plant Sci. 2024. PMID: 38825043
-
Overexpression of the PtrNF-YA6 gene inhibits secondary cell wall thickening in poplar.Plant Sci. 2024 Jun;343:112058. doi: 10.1016/j.plantsci.2024.112058. Epub 2024 Mar 4. Plant Sci. 2024. PMID: 38447913 Review.
-
Xylan in the Middle: Understanding Xylan Biosynthesis and Its Metabolic Dependencies Toward Improving Wood Fiber for Industrial Processing.Front Plant Sci. 2019 Feb 25;10:176. doi: 10.3389/fpls.2019.00176. eCollection 2019. Front Plant Sci. 2019. PMID: 30858858 Free PMC article. Review.
Cited by
-
Deciphering the intricate hierarchical gene regulatory network: unraveling multi-level regulation and modifications driving secondary cell wall formation.Hortic Res. 2023 Dec 19;11(2):uhad281. doi: 10.1093/hr/uhad281. eCollection 2024 Feb. Hortic Res. 2023. PMID: 38344650 Free PMC article.
-
Genome-Wide Identification and Expression Analysis of NF-YA Gene Family in the Filling Stage of Wheat (Triticum aestivum L.).Int J Mol Sci. 2024 Dec 27;26(1):133. doi: 10.3390/ijms26010133. Int J Mol Sci. 2024. PMID: 39795991 Free PMC article.
-
The down-regulation of MsWOX13-2 promotes enhanced waterlogging resilience in alfalfa.Plant J. 2025 Aug;123(4):e70411. doi: 10.1111/tpj.70411. Plant J. 2025. PMID: 40836384 Free PMC article.
-
Comprehensive Analysis of WUSCEL-Related Homeobox Gene Family in Ramie (Boehmeria nivea) Indicates Its Potential Role in Adventitious Root Development.Biology (Basel). 2023 Nov 28;12(12):1475. doi: 10.3390/biology12121475. Biology (Basel). 2023. PMID: 38132301 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

