Lineage and role in integrative taxonomy of a heterotrophic orchid complex
- PMID: 35837745
- PMCID: PMC9452484
- DOI: 10.1111/mec.16617
Lineage and role in integrative taxonomy of a heterotrophic orchid complex
Abstract
Lineage-based species definitions applying coalescent approaches to species delimitation have become increasingly popular. Yet, the application of these methods and the recognition of lineage-only definitions have recently been questioned. Species delimitation criteria that explicitly consider both lineages and evidence for ecological role shifts provide an opportunity to incorporate ecologically meaningful data from multiple sources in studies of species boundaries. Here, such criteria were applied to a problematic group of mycoheterotrophic orchids, the Corallorhiza striata complex, analysing genomic, morphological, phenological, reproductive-mode, niche, and fungal host data. A recently developed method for generating genomic polymorphism data-ISSRseq-demonstrates evidence for four distinct lineages, including a previously unidentified lineage in the Coast Ranges and Cascades of California and Oregon, USA. There is divergence in morphology, phenology, reproductive mode, and fungal associates among the four lineages. Integrative analyses, conducted in population assignment and redundancy analysis frameworks, provide evidence of distinct genomic lineages and a similar pattern of divergence in the extended data, albeit with weaker signal. However, none of the extended data sets fully satisfy the condition of a significant role shift, which requires evidence of fixed differences. The four lineages identified in the current study are recognized at the level of variety, short of comprising different species. This study represents the most comprehensive application of lineage + role to date and illustrates the advantages of such an approach.
Keywords: ISSRseq; Orchidaceae; SNP data; integrative species delimitation; species boundaries; species concept.
© 2022 John Wiley & Sons Ltd.
Figures
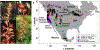




References
-
- Abouheif E (1999). A method for testing the assumption of phylogenetic independence in comparative data. Evolutionary Ecology Research, 1, 895–909.
-
- Ackerfield JR, Keil DJ, Hodgson WC, Simmons Mark. P., Fehlberg SD, & Funk VA (2020). Thistle be a mess: Untangling the taxonomy of Cirsium (Cardueae: Compositae) in North America. Journal of Systematics and Evolution, 58(6), 881–912. 10.1111/jse.12692 - DOI
-
- Ackerman JD (1983). Specificity and mutual dependency of the orchid-euglossine bee interaction. Biological Journal of the Linnean Society, 20(3), 301–314. 10.1111/j.1095-8312.1983.tb01878.x - DOI
-
- Aiello-Lammens ME, Boria RA, Radosavljevic A, Vilela B, & Anderson RP (2015). spThin: An R package for spatial thinning of species occurrence records for use in ecological niche models. Ecography, 38(5), 541–545. 10.1111/ecog.01132 - DOI
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

