Creatine riboside is a cancer cell-derived metabolite associated with arginine auxotrophy
- PMID: 35838048
- PMCID: PMC9282934
- DOI: 10.1172/JCI157410
Creatine riboside is a cancer cell-derived metabolite associated with arginine auxotrophy
Abstract
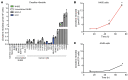
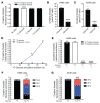
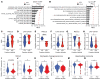
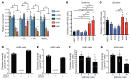
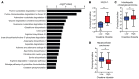
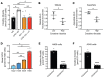
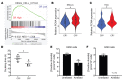
The metabolic dependencies of cancer cells have substantial potential to be exploited to improve the diagnosis and treatment of cancer. Creatine riboside (CR) is identified as a urinary metabolite associated with risk and prognosis in lung and liver cancer. However, the source of high CR levels in patients with cancer as well as their implications for the treatment of these aggressive cancers remain unclear. By integrating multiomics data on lung and liver cancer, we have shown that CR is a cancer cell-derived metabolite. Global metabolomics and gene expression analysis of human tumors and matched liquid biopsies, together with functional studies, revealed that dysregulation of the mitochondrial urea cycle and a nucleotide imbalance were associated with high CR levels and indicators of a poor prognosis. This metabolic phenotype was associated with reduced immune infiltration and supported rapid cancer cell proliferation that drove aggressive tumor growth. CRhi cancer cells were auxotrophic for arginine, revealing a metabolic vulnerability that may be exploited therapeutically. This highlights the potential of CR not only as a poor-prognosis biomarker but also as a companion biomarker to inform the administration of arginine-targeted therapies in precision medicine strategies to improve survival for patients with cancer.
Keywords: Cancer; Lung cancer; Oncology.
Conflict of interest statement
Figures



References
-
- Fendt SM, et al. Targeting metabolic plasticity and flexibility dynamics for cancer therapy. Cancer Discov. 2020;10(12):1797–1807. doi: 10.1158/2159-8290.CD-20-0844. - DOI - PMC - PubMed

